Warfarin PKPD
Nick Holford, Tomoo Funaki
1 Libraries
The first step is to load the libraries we need - in this case, nlmixr and xpose.nlmixr. (We need RxODE and xpose as well, but those are loaded automatically by nlmixr.)
#Parameters
issim=3 # flag for data source: original data (0, simulated data lag time (1), simulated data transit (2) and (3)
ispds=F # flag to use SAEM (T) or FOCEI (F) for EBEs of PK parameters
isplot=0 # flag to show nlmixr diagnostic and residual plots (1) or not (0)
## Getting started
#Advice from nlmixr installation-notes.rtf (nlmixr_1.1.1-3)
#Make sure there is only one library path
.libPaths()
#> [1] "M:/Apps/R/R-4.0.5/library"
.libPaths(.libPaths()[regexpr("nlmixr",.libPaths())!=-1])
srclib=.libPaths() #srclib="M:\\Apps\\R\\R-4.0.5\\library"
## requires patchwork and xpose.nlmixr packages for markdown.
library(ggplot2,lib=srclib)
library(nlmixr,lib=srclib)
library(RxODE,lib=srclib)
#> RxODE 1.0.8 using 3 threads (see ?getRxThreads)
#> no cache: create with `rxCreateCache()`
library(xpose,lib=srclib)
#>
#> Attaching package: 'xpose'
#> The following object is masked from 'package:nlmixr':
#>
#> vpc
#> The following object is masked from 'package:stats':
#>
#> filter
library(xpose.nlmixr,lib=srclib)
#>
#> Attaching package: 'xpose.nlmixr'
#> The following object is masked from 'package:xpose':
#>
#> vpc
library(patchwork,lib=srclib)
#Show versions
packageVersion("vpc")
#> [1] '1.2.2'
packageVersion("RxODE")
#> [1] '1.0.8'
nlmixrver=packageVersion("nlmixr")
nlmixrver
#> [1] '2.0.4'2 Local functions
myvpc=function(nlmixr.fit,ylab) {
nlmixr::vpc(nlmixr.fit,n=500, show=list(obs_dv=T,sim_median=T,pi=T), bins = bintimes,
ylab = ylab, xlab = "Time (hours)", stratify="CMT", scales = "free_y")
}
# Goodness of fit plots using xpose
mygof=function(nlmixr.fit) {
xp.fit <- xpose_data_nlmixr(nlmixr.fit)
dvpred <- dv_vs_pred(xp.fit) + labs(title = NULL, subtitle = NULL, caption = NULL)
dvipred <- dv_vs_ipred(xp.fit) + labs(title = NULL, subtitle = NULL, caption = NULL)
cwresidv <- res_vs_idv(xp.fit) + labs(title = NULL, subtitle = NULL, caption = NULL) # default is CWRES
cwrespred <- res_vs_pred(xp.fit) + labs(title = NULL, subtitle = NULL, caption = NULL) # default is CWRES
return (dvpred + dvipred + cwresidv + cwrespred + plot_layout(nrow=2) + plot_annotation(title=NULL))
}3 Data
Original warfarin PKPD data (O’Reilly et al. 1963, 1968) #O’Reilly RA, Aggeler PM, Leong LS. Studies of the coumarin anticoagulant drugs: The pharmacodynamics of warfarin in man. J Clin Invest. 1963;42(10):1542-51. #O’Reilly RA, Aggeler PM. Studies on coumarin anticoagulant drugs. Initiation of warfarin therapy without a loading dose. Circulation. 1968;38:169-77.
PKPD and turnover model for warfarin. (Holford 1986) #Holford NH. Clinical pharmacokinetics and pharmacodynamics of warfarin. Understanding the dose-effect relationship. Clin Pharmacokinet. 1986;11(6):483-504.
NONMEM PKPD model used to simulate data (Funaki et al. 2018) #Funaki T, Holford N, Fujita S. Population PKPD analysis using nlmixr and NONMEM. Population Approach Group Japan2018.
# Read data
datadir="U:\\Research\\Pharmacometrics\\Users\\Tomoo Funaki\\warfarin\\data"
bintimes=c(seq(0,10,1),seq(12,144,12)) # bins for VPC
if (issim==0) {
datafile="warfarin_dat.csv"
rdata_ext=""
}
if (issim==1) {
datafile="ka1_to_emax1_simln_items.csv"
rdata_ext="_sim"
}
if (issim==2) {
datafile="ka1_to_emax1_simtr_10.csv"
rdata_ext="_simtr"
}
if (issim==3) {
datafile="ka1_to_emax1_simtr2_10.csv"
rdata_ext="_simtr2"
}
if (ispds) {
rdata_ext=paste(rdata_ext,"_PDS",sep="")
} else {
rdata_ext=paste(rdata_ext,"_PDF",sep="")
}
data <- read.csv(file = paste(datadir,datafile,sep="/"))
data$dv <- as.numeric(as.character(data$dv))
names(data) <- toupper(names(data))
data2 <- data
data2$DV <- ifelse(data2$TIME==0 & data2$DVID==1, NA, data2$DV)3.1 Plot data
Plot the PK and INR data
ggplot(data2, aes(x=TIME, y=DV)) +
geom_line(aes(group=ID), col="red") +
scale_x_continuous("Time (h)") +
scale_y_continuous("Concentration | Effect") +
facet_wrap(~DVID, scales = "free_y") +
labs(title="Warfarin single-dose", subtitle="Concentration | Effect vs. time by individual")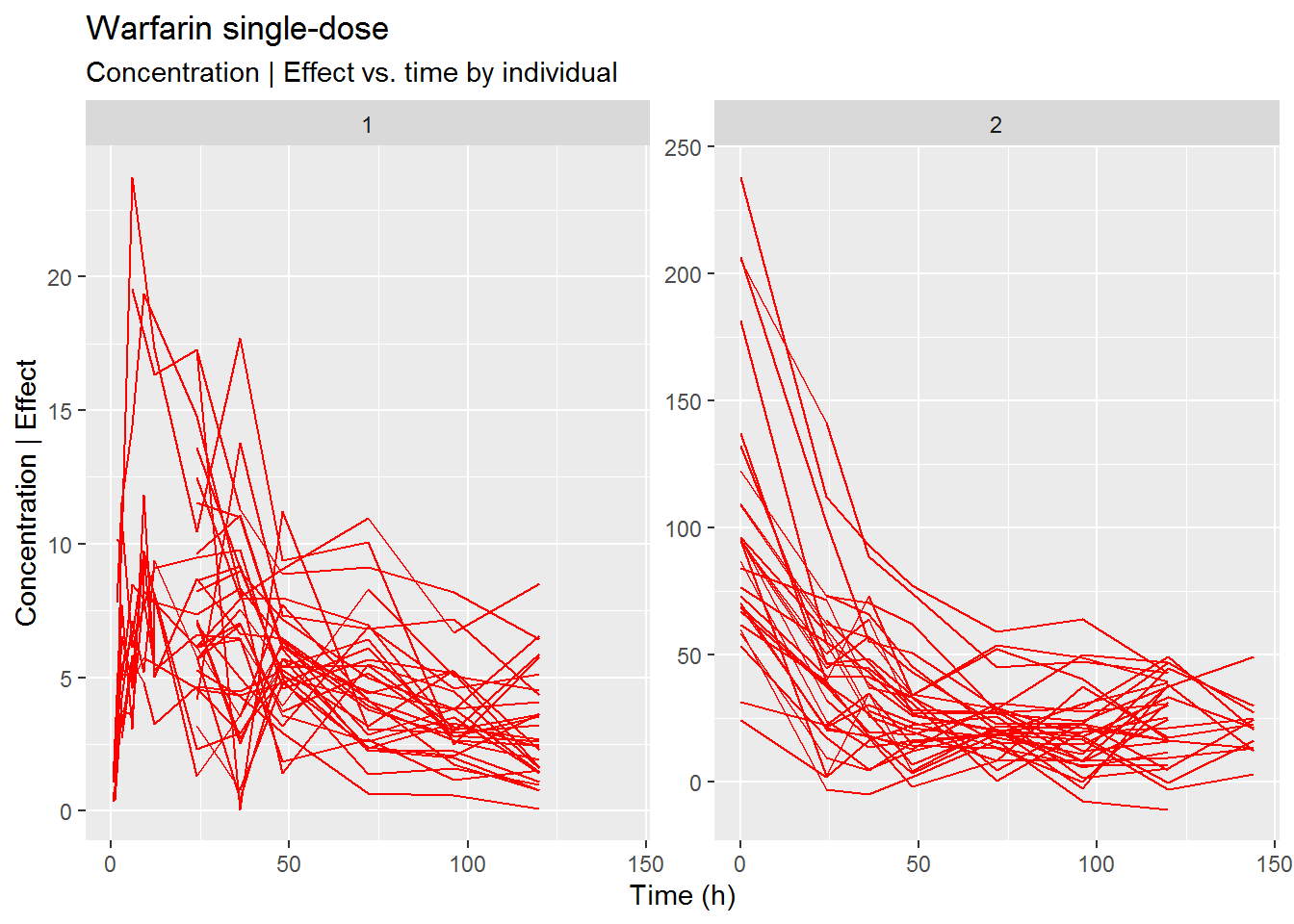
4 PK data
Create PK object from input data
# Selecting only PK data
data.pk <- data[data$DVID==1, ]
data.pk <- data.pk[!is.na(data.pk$DV)|data.pk$AMT!=0, ]
data.pk$CMT <- ifelse(data.pk$AMT!=0, 1, 3)
save(data.pk,file=paste(datadir,"/","datapk",rdata_ext,".RData",sep=""))
#load(paste(datadir,"/",datapk",rdata_ext,".RData",sep=""))5 PK model
5.1 pk.ka1t PK
Let’s start by fitting the PK model using a 1-compartment model with lag time, first-order absorption and elimination.
pk.ka1t <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
eps.pkprop <- 0.1
eps.pkadd <- 0.1
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
cp = center / v
cp ~ prop(eps.pkprop) + add(eps.pkadd)
})
}5.1.1 pk.fit.ka1t SAEM
pk.fit.ka1tS <- nlmixr(pk.ka1t, data.pk, est="saem", control=list(print=0),table=list(cwres=T))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
pk.fit.ka1tS
#> -- nlmixr SAEM(ODE); OBJF by FOCEi approximation fit -------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 613.922 1095.229 1130.484 -537.6145 409.7203
#>
#> -- Time (sec pk.fit.ka1tS$time): ---------------------------------------------------------------------------------------
#>
#> saem setup table cwres covariance other
#> elapsed 32.57 38.226 0.13 38.42 0.09 7.864
#>
#> -- Population Parameters (pk.fit.ka1tS$parFixed or pk.fit.ka1tS$parFixedDf): -------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV%) Shrink(SD)%
#> tktr -0.3 1.14 379 0.741 (0.0798, 6.88) 24.6 78.9%
#> tka -0.119 1.25 1.05e+003 0.888 (0.0765, 10.3) 31.4 76.9%
#> tcl -2.24 0.0891 3.97 0.106 (0.0892, 0.126) 41.2 15.2%
#> tv 2.39 0.087 3.64 10.9 (9.2, 12.9) 43.3 12.1%
#> eps.pkprop 0.349 0.349
#> eps.pkadd 0.19 0.19
#>
#> Covariance Type (pk.fit.ka1tS$covMethod): linFim
#> Some strong fixed parameter correlations exist (pk.fit.ka1tS$cor) :
#> cor:tka,tktr cor:tcl,tktr cor:tv,tktr cor:tcl,tka cor:tv,tka cor:tv,tcl
#> -0.982 -0.0714 0.0580 0.0528 -0.0321 -0.103
#>
#>
#> No correlations in between subject variability (BSV) matrix
#> Full BSV covariance (pk.fit.ka1tS$omega)
#> or correlation ( pk.fit.ka1tS $omegaR ; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#>
#> -- Fit Data (object pk.fit.ka1tS is a modified tibble): ----------------------------------------------------------------
#> # A tibble: 251 x 25
#> ID TIME DV PRED RES WRES IPRED IRES IWRES CPRED CRES CWRES
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.5 0.338 0.576 -0.238 -0.575 0.504 -0.165 -0.639 0.572 -0.234 -0.624
#> 2 1 1 0.784 1.78 -0.996 -0.898 1.58 -0.792 -1.36 1.77 -0.985 -0.994
#> 3 1 2 5.09 4.37 0.717 0.283 3.97 1.12 0.803 4.36 0.732 0.316
#> # ... with 248 more rows, and 13 more variables: eta.ktr <dbl>, eta.ka <dbl>,
#> # eta.cl <dbl>, eta.v <dbl>, depot <dbl>, gut <dbl>, center <dbl>, ktr <dbl>,
#> # ka <dbl>, cl <dbl>, v <dbl>, tad <dbl>, dosenum <dbl>
if (isplot) plot(pk.fit.ka1tS)
#bootstrapFit(pk.fit.ka1tS, nboot = 100, restart = TRUE) # for illustration but this takes a very long time5.1.2 pk.fit.ka1t VPC and GOF
The PK VPC using the original data does not describe the peak very well. Hard to see a problem with standard diagnostics.
vpc=myvpc(pk.fit.ka1tS,ylab="PK Concentration (mg/L) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:03
gof=mygof(pk.fit.ka1tS)
vpc/gof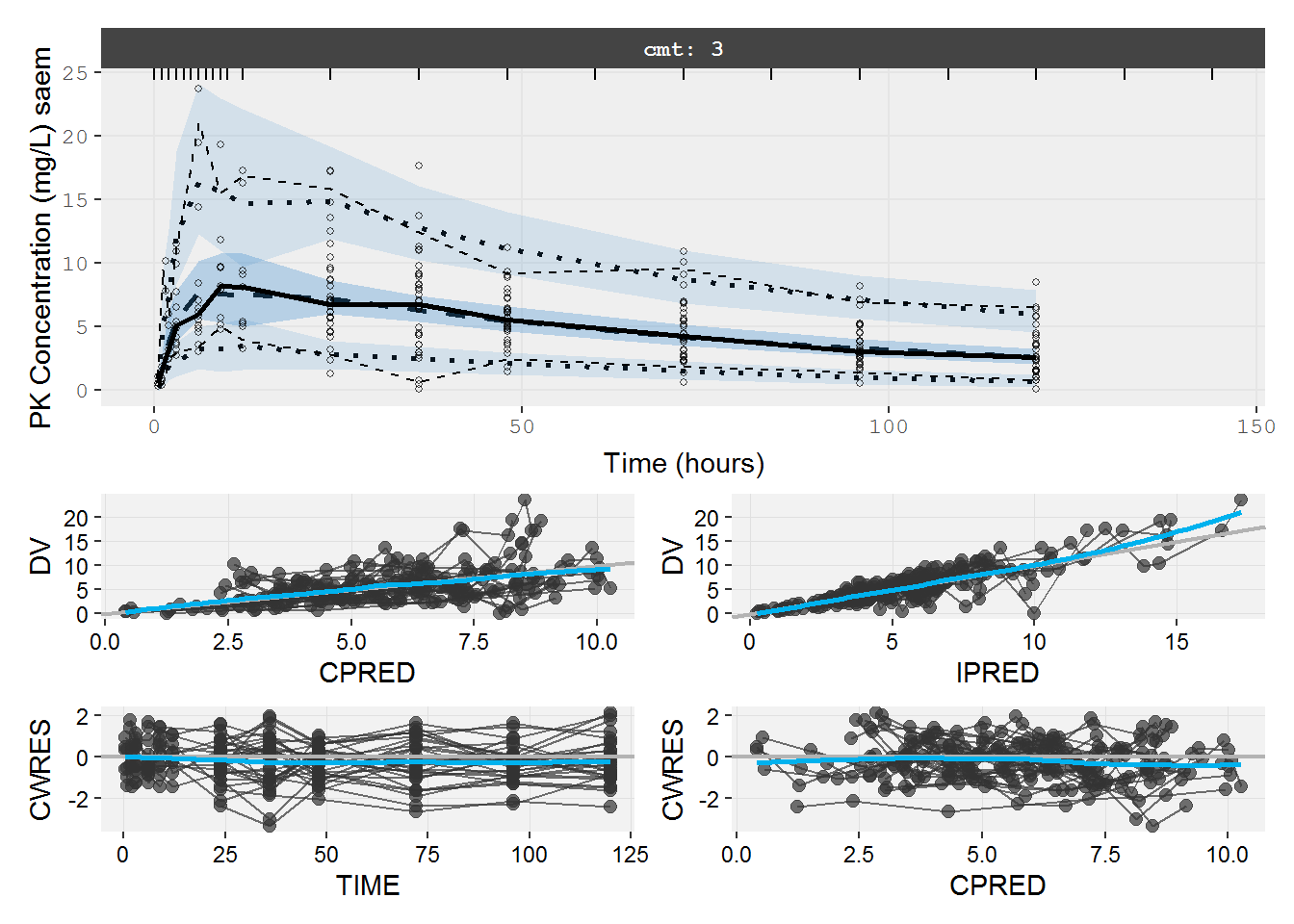
5.1.3 pk.fit.ka1t FOCEI
pk.fit.ka1tF <- nlmixr(pk.ka1t, data.pk, est="focei", control=list(print=0,outerOpt="bobyqa",table=list(cwres=T)))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:00:11
#> done
pk.fit.ka1tF
#> -- nlmixr FOCEi (outer: bobyqa) fit ------------------------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 606.1466 1087.454 1122.708 -533.7269 41.21103
#>
#> -- Time (sec pk.fit.ka1tF$time): ---------------------------------------------------------------------------------------
#>
#> setup optimize covariance table other
#> elapsed 55.284 11.828 11.828 0.08 33.74
#>
#> -- Population Parameters (pk.fit.ka1tF$parFixed or pk.fit.ka1tF$parFixedDf): -------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV%) Shrink(SD)%
#> tktr 0.267 0.128 47.9 1.31 (1.02, 1.68) 10.0 89.6%
#> tka -0.67 0.089 13.3 0.512 (0.43, 0.609) 10.1 84.4%
#> tcl -2.2 0.0224 1.02 0.11 (0.106, 0.115) 42.9 13.2%
#> tv 2.39 0.0695 2.9 10.9 (9.54, 12.5) 46.3 8.19%
#> eps.pkprop 0.332 0.332
#> eps.pkadd 0.144 0.144
#>
#> Covariance Type (pk.fit.ka1tF$covMethod): r,s
#> Fixed parameter correlations in pk.fit.ka1tF$cor
#> No correlations in between subject variability (BSV) matrix
#> Full BSV covariance (pk.fit.ka1tF$omega)
#> or correlation ( pk.fit.ka1tF $omegaR ; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#> Minimization message (pk.fit.ka1tF$message):
#> Normal exit from bobyqa
#>
#> -- Fit Data (object pk.fit.ka1tF is a modified tibble): ----------------------------------------------------------------
#> # A tibble: 251 x 25
#> ID TIME DV PRED RES WRES IPRED IRES IWRES CPRED CRES CWRES
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.5 0.338 0.569 -0.230 -0.655 0.535 -0.197 -0.862 0.568 -0.229 -0.686
#> 2 1 1 0.784 1.72 -0.937 -0.961 1.63 -0.843 -1.51 1.72 -0.934 -1.01
#> 3 1 2 5.09 4.14 0.952 0.415 3.95 1.15 0.870 4.13 0.956 0.437
#> # ... with 248 more rows, and 13 more variables: eta.ktr <dbl>, eta.ka <dbl>,
#> # eta.cl <dbl>, eta.v <dbl>, depot <dbl>, gut <dbl>, center <dbl>, ktr <dbl>,
#> # ka <dbl>, cl <dbl>, v <dbl>, tad <dbl>, dosenum <dbl>
if (isplot) plot(pk.fit.ka1tF)5.1.4 pk.fit.ka1t VPC & GOF
The PK VPC using the original data does not describe the peak very well. Hard to see a problem with standard diagnostics.
vpc=myvpc(pk.fit.ka1tF,ylab="PK Concentration (mg/L) focei")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
gof=mygof(pk.fit.ka1tF)
vpc/gof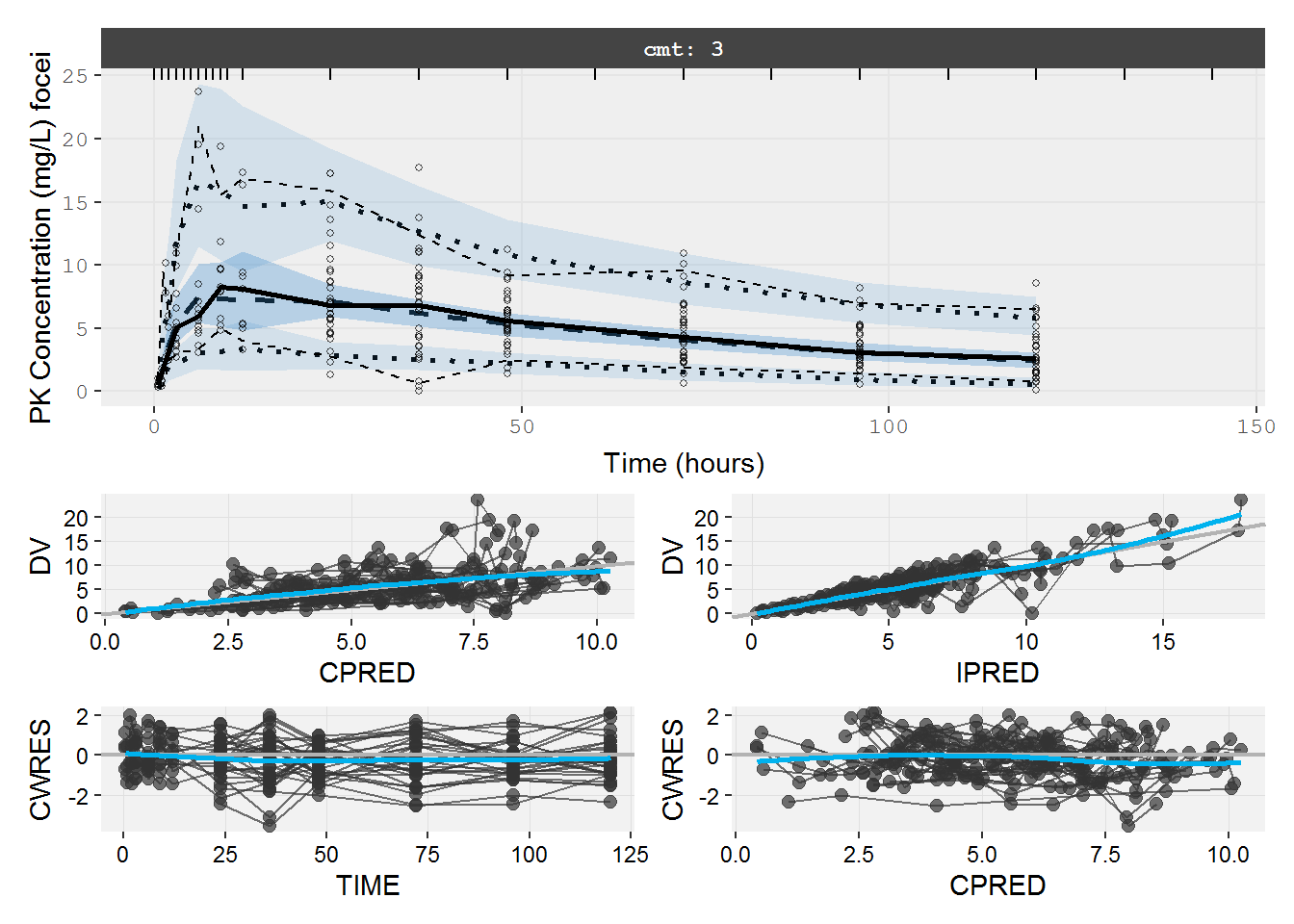
6 PD data
6.1 PD data object with PK EBEs
The PK model empirical Bayes estimates are combined with the effect data to allow a sequential PKPD model.
# Make PD data with SAEM (ispds=1) or FOCEI (ispds=0) EBEs for PK parameters
if (ispds) {
fit.ipara <- pk.fit.ka1tS[pk.fit.ka1tS$TIME==24,]
} else {
}
#> NULL
fit.ipara <- pk.fit.ka1tF[pk.fit.ka1tF$TIME==24,]
fit.ipara <- fit.ipara[c("ID", "ktr", "ka", "cl", "v")]
names(fit.ipara) <- c("ID", "IKTR", "IKA", "ICL", "IV")
data.pd <- data
data.pd <- merge(data.pd, fit.ipara, by="ID", all=T)
data.pd$CMT <- ifelse(data.pd$AMT!=0, 1,
ifelse(data.pd$DVID==2 & data.pd$AMT==0, 4, 3))
#Create MDV=1 for dose record or when DV is recorded as zero
data.pd$MDV <- ifelse(data.pd$AMT>0 | data.pd$DV==0, 1, 0)
#select dosing records and PCA effect records
data.pd <- data.pd[data.pd$AMT!=0 | data.pd$DVID==2, ]
save(data.pd,file=paste(datadir,"/","datapd",rdata_ext,".RData",sep=""))
#load(file=paste(datadir,"/","datapd",rdata_ext,".RData",sep=""))6.2 PKPD data object
The PKPD data object includes both concentration and effect observations.
data.pkpd <- read.csv(file = paste(datadir,datafile,sep="/"))
names(data.pkpd) = toupper(names(data.pkpd))
data.pkpd$CMT <- data.pkpd$DVID + 2 # DVID 0->2 (center);DVID 2->3 (center); DVID 3->4 (effect)
data.pkpd$CMT[data.pkpd$AMT != 0] <- 1 # CMT 2->1
data.pkpd$EVID[data.pkpd$AMT != 0] <- 101
save(data.pkpd,file=paste(datadir,"/","datapkpd",rdata_ext,".RData",sep=""))
#load(file=paste(datadir,"/","datapkpd",rdata_ext,".RData",sep=""))7 Sequential PD Models
7.1 pd.cp.logit Immediate PD logit
Sequential immediate effect model - logit for Emax
# define the models
pd.cp.logit <- function() {
ini({
poplogit <- 7.5
tc50 <- log(0.5)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
e0 = exp(te0 + eta.e0)
# PK parameters from input dataset
ktr = IKTR
ka = IKA
cl = ICL
v = IV
cp = center/v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
effect=e0*(1-emax*cp/(c50+cp))
effect ~ add(eps.pdadd)
})
}7.1.1 pd.fit.CPS SAEM
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "IKTR" "IKA" "ICL" "IV"
#> -- nlmixr SAEM(ODE); OBJF by FOCEi approximation fit -------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 1689.296 2129.683 2153.81 -1057.842 6.406401
#>
#> -- Time (sec pd.fit.CPS$time): -----------------------------------------------------------------------------------------
#>
#> saem setup table cwres covariance other
#> elapsed 40.78 45.655 0.08 45.81 0.06 6.055
#>
#> -- Population Parameters (pd.fit.CPS$parFixed or pd.fit.CPS$parFixedDf): -----------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV% or SD) Shrink(SD)%
#> poplogit 1.4 0.144 10.3 1.4 (1.12, 1.69) 0.485 49.2%
#> tc50 -0.547 0.0574 10.5 0.579 (0.517, 0.648) 20.4 82.1%
#> te0 4.47 0.0816 1.82 87.3 (74.4, 102) 43.3 9.31%
#> eps.pdadd 19.8 19.8
#>
#> Covariance Type (pd.fit.CPS$covMethod): linFim
#> Fixed parameter correlations in pd.fit.CPS$cor
#> No correlations in between subject variability (BSV) matrix
#> Full BSV covariance (pd.fit.CPS$omega)
#> or correlation ( pd.fit.CPS $omegaR ; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#>
#> -- Fit Data (object pd.fit.CPS is a modified tibble): ------------------------------------------------------------------
#> # A tibble: 232 x 33
#> ID TIME DV PRED RES WRES IPRED IRES IWRES CPRED CRES CWRES
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 24 73.5 22.5 51.0 2.24 24.4 49.0 2.47 22.4 51.1 2.19
#> 2 1 36 66.0 23.1 42.9 1.87 25.0 41.0 2.07 23.1 42.9 1.84
#> 3 1 48 45.4 23.9 21.5 0.935 25.7 19.7 0.992 23.8 21.6 0.919
#> # ... with 229 more rows, and 21 more variables: eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>, logit <dbl>,
#> # emax <dbl>, c50 <dbl>, e0 <dbl>, ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>,
#> # cp <dbl>, tad <dbl>, dosenum <dbl>, IV <dbl>, ICL <dbl>, IKA <dbl>,
#> # IKTR <dbl>7.1.2 pd.fit.CPS VPC & GOF
The VPC and GOF plots shows the immediate effect model describes the data poorly.
vpc=myvpc(pd.fit.CPS,ylab="CPS Response (PCA) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:03
gof=mygof(pd.fit.CPS)
vpc/gof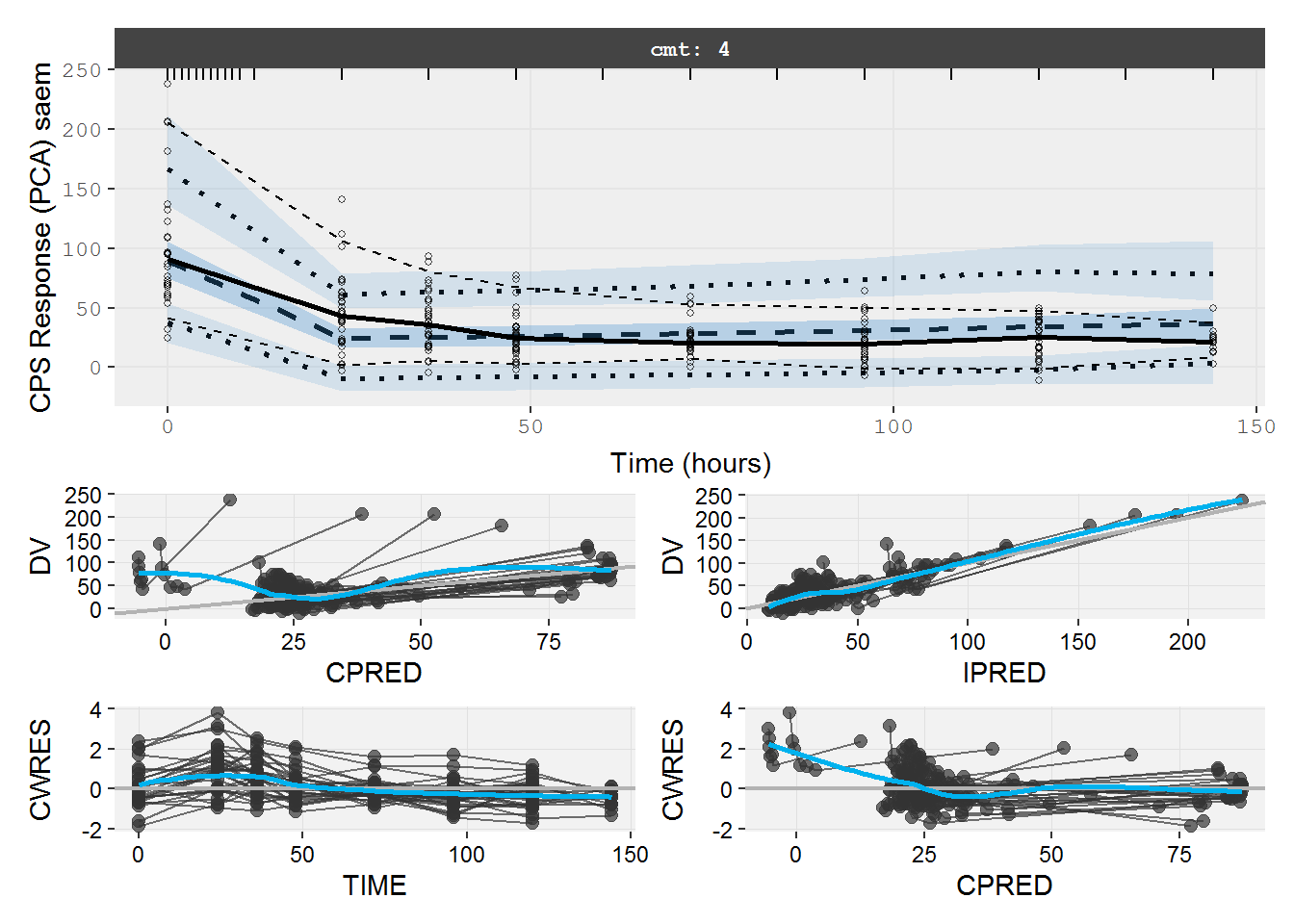
7.2 pd.ce.logit Effect CMT PD logit
Sequential effect compartment model - logit for Emax
pd.ce.logit <- function() {
ini({
poplogit <- 7.5
tc50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
# PK parameters from input dataset
ktr = IKTR
ka = IKA
cl = ICL
v = IV
cp = center/v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
d/dt(ce) = kout*(cp - ce)
effect=e0*(1-emax*ce/(c50+ce))
effect ~ add(eps.pdadd)
})
}7.2.1 pd.fit.CES SAEM
pd.fit.CES <- nlmixr(pd.ce.logit, data.pd, est="saem", control=list(print=0),table=list(cwres=T))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "IKTR" "IKA" "ICL" "IV"
if (isplot) plot(pd.fit.CES)
pd.fit.CES
#> -- nlmixr SAEM(ODE); OBJF by FOCEi approximation fit -------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 1534.141 1978.528 2009.549 -980.264 573.5914
#>
#> -- Time (sec pd.fit.CES$time): -----------------------------------------------------------------------------------------
#>
#> saem setup table cwres covariance other
#> elapsed 75.11 47.71 0.1 47.9 0.08 5.93
#>
#> -- Population Parameters (pd.fit.CES$parFixed or pd.fit.CES$parFixedDf): -----------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV% or SD)
#> poplogit 9.85 1.46 14.9 9.85 (6.98, 12.7) 1.63
#> tc50 -0.676 0.0738 10.9 0.508 (0.44, 0.587) 18.3
#> tkout -5.53 0.146 2.64 0.00397 (0.00298, 0.00528) 51.1
#> te0 4.51 0.0833 1.85 90.9 (77.2, 107) 45.9
#> eps.pdadd 12.7 12.7
#> Shrink(SD)%
#> poplogit 91.8%
#> tc50 67.9%
#> tkout 29.6%
#> te0 4.07%
#> eps.pdadd
#>
#> Covariance Type (pd.fit.CES$covMethod): linFim
#> Fixed parameter correlations in pd.fit.CES$cor
#> No correlations in between subject variability (BSV) matrix
#> Full BSV covariance (pd.fit.CES$omega)
#> or correlation ( pd.fit.CES $omegaR ; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#>
#> -- Fit Data (object pd.fit.CES is a modified tibble): ------------------------------------------------------------------
#> # A tibble: 232 x 36
#> ID TIME DV PRED RES WRES IPRED IRES IWRES CPRED CRES CWRES
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 24 73.5 40.2 33.3 1.37 54.9 18.6 1.46 37.8 35.7 1.16
#> 2 1 36 66.0 32.3 33.7 1.57 44.3 21.7 1.70 30.3 35.7 1.33
#> 3 1 48 45.4 27.8 17.6 0.887 38.2 7.14 0.561 26.1 19.3 0.792
#> # ... with 229 more rows, and 24 more variables: eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.kout <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>,
#> # ce <dbl>, logit <dbl>, emax <dbl>, c50 <dbl>, kout <dbl>, e0 <dbl>,
#> # ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>, cp <dbl>, tad <dbl>, dosenum <dbl>,
#> # IV <dbl>, ICL <dbl>, IKA <dbl>, IKTR <dbl>7.2.2 pd.fit.CES VPC & GOF
The VPC and GOF show the effect compartment model does not describe the data well.
vpc=myvpc(pd.fit.CES,ylab="CES Response (PCA) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
gof=mygof(pd.fit.CES)
vpc/gof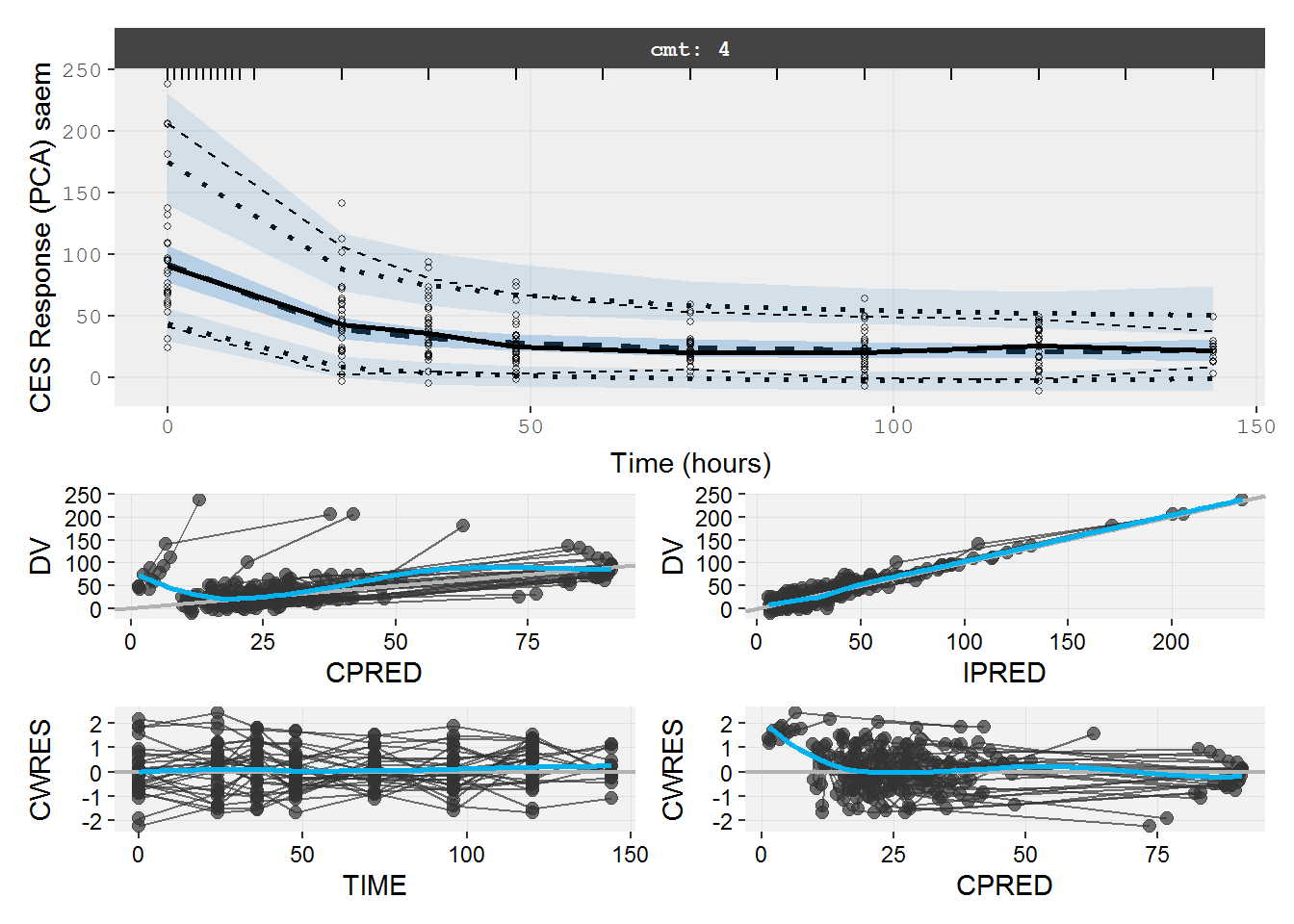
7.3 pd.turnover.logit Turnover PD logit
Sequential turnover model - logit for Emax
pd.turnover.logit <- function() { # The logit transformation is used to constrain Emax in SAEM
ini({
poplogit <- 7.5
tc50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
# PK parameters from input dataset
ktr = IKTR
ka = IKA
cl = ICL
v = IV
cp = center/v
PD=1-emax*cp/(c50+cp)
effect(0) = e0
kin = e0*kout
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
d/dt(effect) = kin*PD -kout*effect
effect ~ add(eps.pdadd)
})
}7.3.1 pd.fit.TOS SAEM
Sequential turnover model logit
################ Run nlmixr with PD and PKPD models ###########################
pd.fit.TOS <- nlmixr(pd.turnover.logit, data.pd, est="saem", control=list(print=0),table=list(cwres=T))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "IKTR" "IKA" "ICL" "IV"
if (isplot) plot(pd.fit.TOS)
pd.fit.TOS
#> -- nlmixr SAEM(ODE); OBJF by FOCEi approximation fit -------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 1507.262 1951.649 1982.67 -966.8246 48.28872
#>
#> -- Time (sec pd.fit.TOS$time): -----------------------------------------------------------------------------------------
#>
#> saem setup table cwres covariance other
#> elapsed 62.79 47.356 0.13 47.65 0.06 6.564
#>
#> -- Population Parameters (pd.fit.TOS$parFixed or pd.fit.TOS$parFixedDf): -----------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV% or SD) Shrink(SD)%
#> poplogit 7.53 0.581 7.72 7.53 (6.39, 8.67) 1.55 90.6%
#> tc50 -0.302 0.112 37 0.74 (0.594, 0.921) 48.0 36.2%
#> tkout -3.31 0.109 3.3 0.0365 (0.0295, 0.0452) 53.0 18.1%
#> te0 4.5 0.0839 1.86 90.4 (76.7, 106) 48.0 3.14%
#> eps.pdadd 10.9 10.9
#>
#> Covariance Type (pd.fit.TOS$covMethod): linFim
#> Fixed parameter correlations in pd.fit.TOS$cor
#> No correlations in between subject variability (BSV) matrix
#> Full BSV covariance (pd.fit.TOS$omega)
#> or correlation ( pd.fit.TOS $omegaR ; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#>
#> -- Fit Data (object pd.fit.TOS is a modified tibble): ------------------------------------------------------------------
#> # A tibble: 232 x 38
#> ID TIME DV PRED RES WRES IPRED IRES IWRES CPRED CRES CWRES
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 24 73.5 43.3 30.2 1.11 70.0 3.51 0.322 37.0 36.5 0.938
#> 2 1 36 66.0 31.2 34.8 1.51 53.1 12.9 1.19 24.7 41.3 1.21
#> 3 1 48 45.4 23.7 21.6 1.09 41.0 4.35 0.399 17.4 28.0 0.934
#> # ... with 229 more rows, and 26 more variables: eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.kout <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>,
#> # effect <dbl>, logit <dbl>, emax <dbl>, c50 <dbl>, kout <dbl>, e0 <dbl>,
#> # ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>, cp <dbl>, PD <dbl>, kin <dbl>,
#> # tad <dbl>, dosenum <dbl>, IV <dbl>, ICL <dbl>, IKA <dbl>, IKTR <dbl>7.3.2 pd.fit.TOS VPC & GOF
The VPC and GOF show the turnover model describes the PD data well.
vpc=myvpc(pd.fit.TOS,ylab="pd TOS Response (PCA) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:03
gof=mygof(pd.fit.TOS)
vpc/gof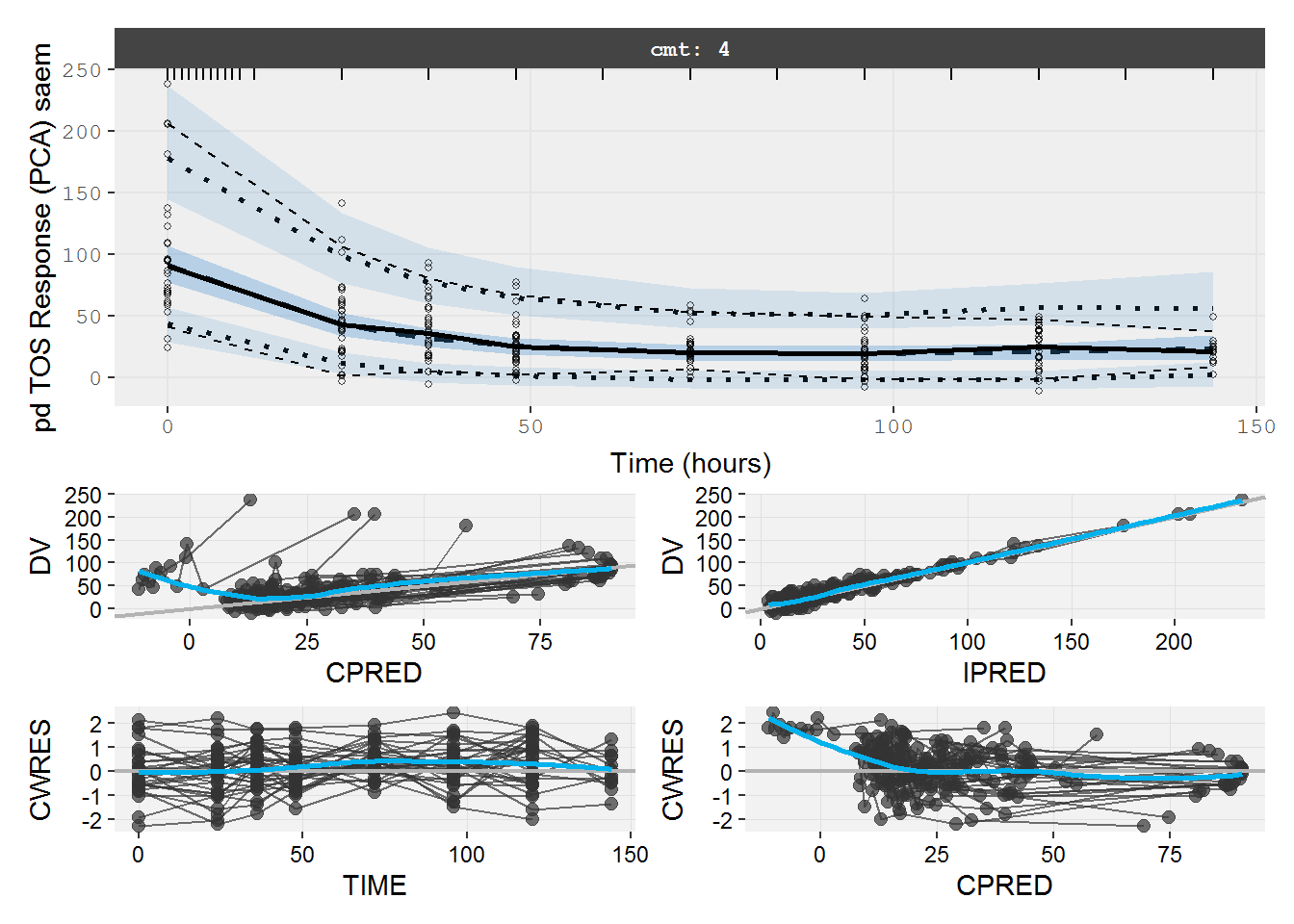
7.4 pd.cp.emax Immediate PD Emax
Sequential immediate effect model Emax<1
pd.cp.emax <- function() {
ini({
popemax <- c(0,0.9,0.999)
tc50 <- log(0.5)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
poplogit = log(popemax/(1-popemax))
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
e0 = exp(te0 + eta.e0)
# PK parameters from input dataset
ktr = IKTR
ka = IKA
cl = ICL
v = IV
cp = center/v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
effect=e0*(1-emax*cp/(c50+cp))
effect ~ add(eps.pdadd)
})
}7.4.1 pd.fit.CPF FOCEI
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "IKTR" "IKA" "ICL" "IV"
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:00:05
#> done
#> -- nlmixr FOCEi (outer: bobyqa) fit ------------------------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 1646.729 2087.117 2111.244 -1036.558 48709
#>
#> -- Time (sec pd.fit.CPF$time): -----------------------------------------------------------------------------------------
#>
#> setup optimize covariance table other
#> elapsed 42.788 5.337 5.337 0.11 58.588
#>
#> -- Population Parameters (pd.fit.CPF$parFixed or pd.fit.CPF$parFixedDf): -----------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV%) Shrink(SD)%
#> popemax 0.707 0.0308 4.36 0.707 (0.646, 0.767)
#> tc50 -6.43 6.62 103 0.00162 (3.79e-009, 693) 320. 99.2%
#> te0 4.53 0.116 2.56 92.4 (73.6, 116) 43.2 9.26%
#> eps.pdadd 17.5 17.5
#>
#> -- BSV Covariance (pd.fit.CPF$omega): ----------------------------------------------------------------------------------
#> eta.emax eta.c50 eta.e0
#> eta.emax 0.1362351 0.000000 0.0000000
#> eta.c50 0.0000000 2.422088 0.0000000
#> eta.e0 0.0000000 0.000000 0.1710548
#>
#> Not all variables are mu-referenced.
#> Can also see BSV Correlation (pd.fit.CPF$omegaR; diagonals=SDs)
#> Covariance Type (pd.fit.CPF$covMethod): r
#> Fixed parameter correlations in pd.fit.CPF$cor
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#> Minimization message (pd.fit.CPF$message):
#> Normal exit from bobyqa
#>
#> -- Fit Data (object pd.fit.CPF is a modified tibble): ------------------------------------------------------------------
#> # A tibble: 232 x 34
#> ID TIME DV PRED RES WRES IPRED IRES IWRES CPRED CRES CWRES
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 24 73.5 27.1 46.4 2.11 32.7 40.8 2.33 26.6 46.9 1.99
#> 2 1 36 66.0 27.1 38.9 1.77 32.7 33.3 1.91 26.6 39.4 1.67
#> 3 1 48 45.4 27.1 18.2 0.832 32.7 12.7 0.725 26.6 18.8 0.794
#> # ... with 229 more rows, and 22 more variables: eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>, poplogit <dbl>,
#> # logit <dbl>, emax <dbl>, c50 <dbl>, e0 <dbl>, ktr <dbl>, ka <dbl>,
#> # cl <dbl>, v <dbl>, cp <dbl>, tad <dbl>, dosenum <dbl>, IV <dbl>, ICL <dbl>,
#> # IKA <dbl>, IKTR <dbl>7.4.2 pd.fit.CPF VPC & GOF
The VPC and GOF shows the immediate effect model does not describe the PD data well.
vpc=myvpc(pd.fit.CPF,ylab="pd CPF Response (PCA) focei")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
gof=mygof(pd.fit.CPF)
vpc/gof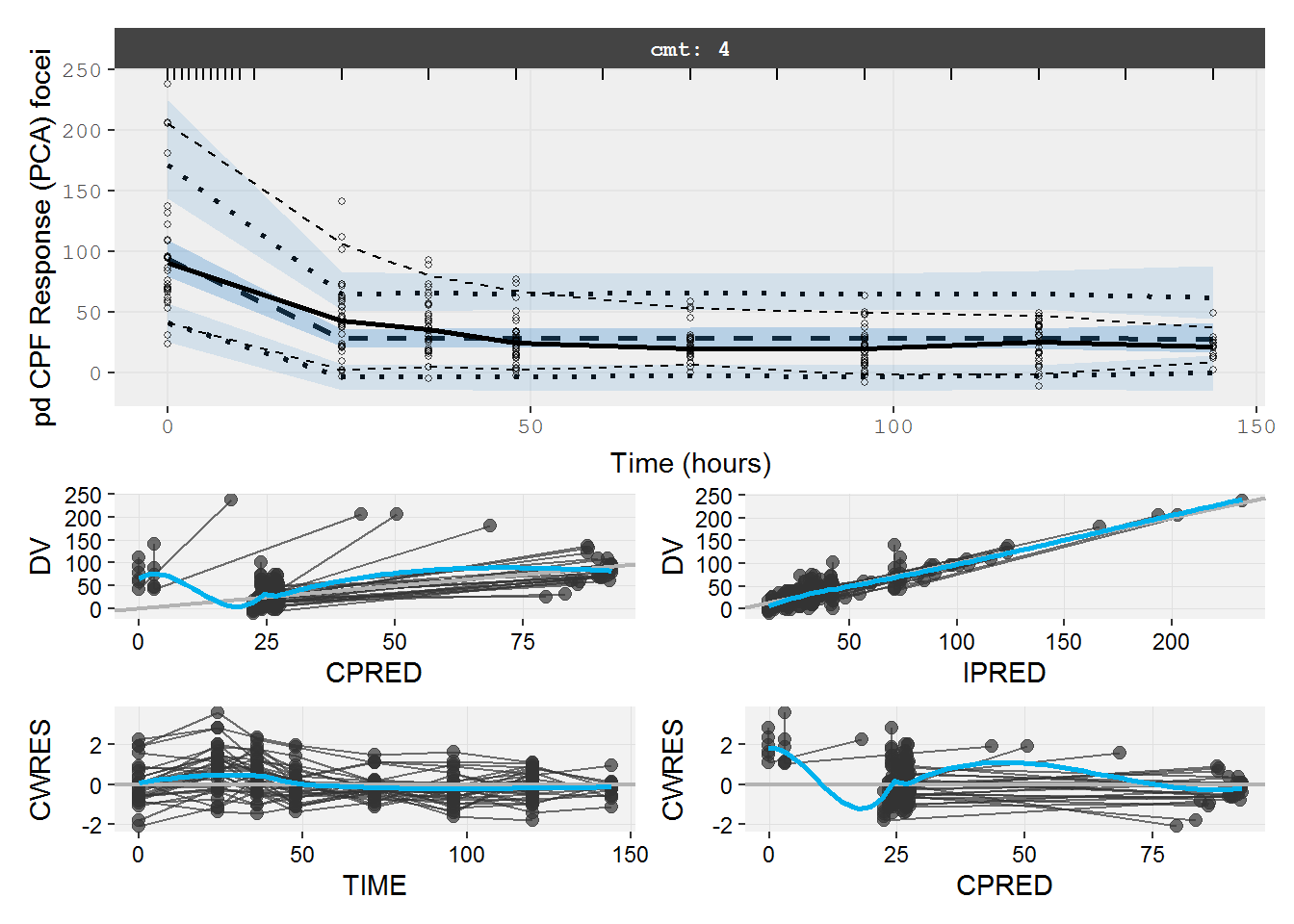
7.5 pd.ce.emax effect CMT Emax
Sequential effect compartment model Emax<1
pd.ce.emax <- function() { #clearer emax parameter constraint method which works with FOCEI
ini({
popemax <- c(0,0.9,0.999)
tc50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
poplogit = log(popemax/(1-popemax))
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
# PK parameters from input dataset
ktr = IKTR
ka = IKA
cl = ICL
v = IV
cp = center/v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
d/dt(ce) = kout*(cp - ce)
effect=e0*(1-emax*ce/(c50+ce))
effect ~ add(eps.pdadd)
})
}7.5.1 pd.fit.CEF FOCEI
################ Effect compartment PD model FOCEI ###########
# pd.fit.CEF <- nlmixr(pd.ce.emax, data.pd, est="focei", control=foceiControl(print=100000)) # Default opt method fails
pd.fit.CEF <- nlmixr(pd.ce.emax, data.pd, est="focei", control=list(print=0,outerOpt="bobyqa",table=list(cwres=T)))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "IKTR" "IKA" "ICL" "IV"
#> unhandled error message: EE:lsoda -- at t = 8.30738e-009 and step size _C(h) = 3.16901e-014, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|===
S matrix calculation failed; Switch to R-matrix covariance.
#> =|====] 0:00:08
#> done
if (isplot) plot(pd.fit.CEF)
pd.fit.CEF
#> -- nlmixr FOCEi (outer: bobyqa) fit ------------------------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood
#> FOCEi 5e+100 5e+100 5e+100 -2.5e+100
#>
#> -- Time (sec pd.fit.CEF$time): -----------------------------------------------------------------------------------------
#>
#> setup optimize covariance table other
#> elapsed 46.547 8.448 8.448 0.15 12.657
#>
#> -- Population Parameters (pd.fit.CEF$parFixed or pd.fit.CEF$parFixedDf): -----------------------------------------------
#>
#> Est. Back-transformed BSV(CV%) Shrink(SD)%
#> popemax 0.836 0.836
#> tc50 -0.702 0.496 76.4 72.2%
#> tkout -3 0.0496 79.7 -534.%
#> te0 4.5 89.7 43.3 16.1%
#> eps.pdadd 22 22
#>
#> -- BSV Covariance (pd.fit.CEF$omega): ----------------------------------------------------------------------------------
#> eta.emax eta.c50 eta.kout eta.e0
#> eta.emax 0.3979735 0.0000000 0.0000000 0.0000000
#> eta.c50 0.0000000 0.4600564 0.0000000 0.0000000
#> eta.kout 0.0000000 0.0000000 0.4922557 0.0000000
#> eta.e0 0.0000000 0.0000000 0.0000000 0.1718628
#>
#> Not all variables are mu-referenced.
#> Can also see BSV Correlation (pd.fit.CEF$omegaR; diagonals=SDs)
#> Covariance Type (pd.fit.CEF$covMethod): failed
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#> Minimization message (pd.fit.CEF$message):
#> Normal exit from bobyqa
#>
#> -- Fit Data (object pd.fit.CEF is a modified tibble): ------------------------------------------------------------------
#> # A tibble: 232 x 37
#> ID TIME DV PRED RES WRES IPRED IRES IWRES CPRED CRES CWRES
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 24 73.5 21.4 52.1 2.07 21.4 52.1 2.37 21.4 52.1 2.07
#> 2 1 36 66.0 20.6 45.4 1.81 20.6 45.4 2.06 20.6 45.4 1.81
#> 3 1 48 45.4 20.6 24.8 0.992 20.6 24.8 1.12 20.6 24.8 0.992
#> # ... with 229 more rows, and 25 more variables: eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.kout <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>,
#> # ce <dbl>, poplogit <dbl>, logit <dbl>, emax <dbl>, c50 <dbl>, kout <dbl>,
#> # e0 <dbl>, ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>, cp <dbl>, tad <dbl>,
#> # dosenum <dbl>, IV <dbl>, ICL <dbl>, IKA <dbl>, IKTR <dbl>7.5.2 pd.fit.CEF VPC & GOF
vpc=myvpc(pd.fit.CEF,ylab="pd CEF Response (PCA) focei")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "IKTR" "IKA" "ICL" "IV"
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#> [====|====|====|====|====|====|====|====|====|====
gof=mygof(pd.fit.CEF)
vpc/gof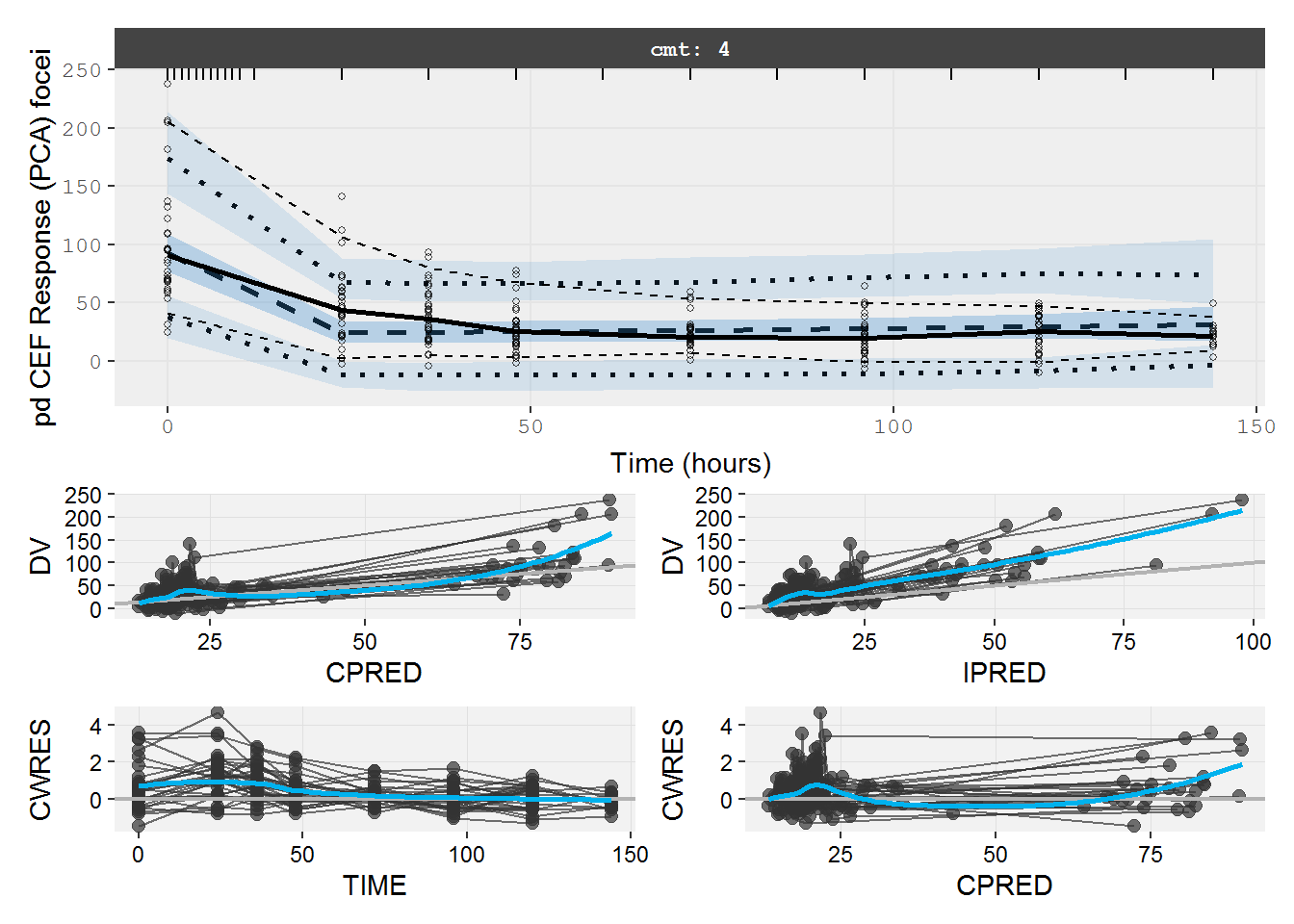
7.6 pd.turnover.emax Turnover PD Emax
Sequential turnover model Emax<1
pd.turnover.emax <- function() {
ini({
popemax <- c(0,0.9,0.999)
tc50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
poplogit = log(popemax/(1-popemax))
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
# PK parameters from input dataset
ktr = IKTR
ka = IKA
cl = ICL
v = IV
cp = center/v
PD=1-emax*cp/(c50+cp)
effect(0) = e0
kin = e0*kout
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
d/dt(effect) = kin*PD -kout*effect
effect ~ add(eps.pdadd)
})
}7.6.1 pd.fit.TOF FOCEI
################ Turnover (true) PD model FOCEI ###########
pd.fit.TOF <- nlmixr(pd.turnover.emax, data.pd, est="focei", control=list(print=0,outerOpt="bobyqa",table=list(cwres=T)))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "IKTR" "IKA" "ICL" "IV"
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:00:27
#> done
if (isplot) plot(pd.fit.TOF)
pd.fit.TOF
#> -- nlmixr FOCEi (outer: bobyqa) fit ------------------------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 1507.147 1951.534 1982.555 -966.7672 296.9303
#>
#> -- Time (sec pd.fit.TOF$time): -----------------------------------------------------------------------------------------
#>
#> setup optimize covariance table other
#> elapsed 55.823 27.571 27.571 0.16 74.245
#>
#> -- Population Parameters (pd.fit.TOF$parFixed or pd.fit.TOF$parFixedDf): -----------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV%) Shrink(SD)%
#> popemax 0.925 0.0208 2.25 0.925 (0.884, 0.965)
#> tc50 -0.773 0.234 30.3 0.462 (0.292, 0.73) 46.1 52.2%
#> tkout -3.21 0.105 3.28 0.0404 (0.0329, 0.0497) 58.2 23.0%
#> te0 4.51 0.0737 1.64 90.5 (78.3, 105) 49.0 7.50%
#> eps.pdadd 10.8 10.8
#>
#> -- BSV Covariance (pd.fit.TOF$omega): ----------------------------------------------------------------------------------
#> eta.emax eta.c50 eta.kout eta.e0
#> eta.emax 0.5424419 0.0000000 0.0000000 0.0000000
#> eta.c50 0.0000000 0.1927781 0.0000000 0.0000000
#> eta.kout 0.0000000 0.0000000 0.2913137 0.0000000
#> eta.e0 0.0000000 0.0000000 0.0000000 0.2151494
#>
#> Not all variables are mu-referenced.
#> Can also see BSV Correlation (pd.fit.TOF$omegaR; diagonals=SDs)
#> Covariance Type (pd.fit.TOF$covMethod): r,s
#> Fixed parameter correlations in pd.fit.TOF$cor
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#> Minimization message (pd.fit.TOF$message):
#> Normal exit from bobyqa
#>
#> -- Fit Data (object pd.fit.TOF is a modified tibble): ------------------------------------------------------------------
#> # A tibble: 232 x 39
#> ID TIME DV PRED RES WRES IPRED IRES IWRES CPRED CRES CWRES
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 24 73.5 42.3 31.2 1.13 70.7 2.78 0.256 34.8 38.7 0.950
#> 2 1 36 66.0 30.7 35.3 1.51 53.7 12.4 1.14 23.2 42.9 1.20
#> 3 1 48 45.4 23.9 21.5 1.08 41.7 3.67 0.338 16.6 28.8 0.919
#> # ... with 229 more rows, and 27 more variables: eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.kout <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>,
#> # effect <dbl>, poplogit <dbl>, logit <dbl>, emax <dbl>, c50 <dbl>,
#> # kout <dbl>, e0 <dbl>, ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>, cp <dbl>,
#> # PD <dbl>, kin <dbl>, tad <dbl>, dosenum <dbl>, IV <dbl>, ICL <dbl>,
#> # IKA <dbl>, IKTR <dbl>7.6.2 pd.fit.TOF VPC & GOF
Quitting from lines 704-711 (warf_simtr_PKPD_vpc_ui_saem_foce_B-18-v3_PDF.Rmd) Error in foceiFit.data.frame0(data = .dat, inits = .inits, PKpars = .uif$theta.pars, : Could not fit data. Calls:
#vpc=myvpc(pd.fit.TOF,ylab="pd TOF Response (PCA) focei")
#gof=mygof(pd.fit.TOF)
#vpc/gof8 Joint PKPD models
We can jointly fit the PK and PD data with a single model.
8.1 pkpd.cp.logit Imm PKPD logit
Joint turnover model Emax<1
pkpd.cp.logit <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
eps.pkprop <- 0.1
eps.pkadd <- 0.1
poplogit <- 7.5
tc50 <- log(0.5)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
e0 = exp(te0 + eta.e0)
cp = center / v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl * cp
effect=e0*(1-emax*cp/(c50+cp))
cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
effect ~ add(eps.pdadd) | ce
})
}8.1.1 pkpd.fit.CPS SAEM
#> __ RxODE-based ODE model _______________________________________________________________________________________________
#> -- Initialization: -----------------------------------------------------------------------------------------------------
#> Fixed Effects ($theta):
#> tktr tka tcl tv poplogit tc50 te0
#> 0.0000000 0.0000000 -2.3025851 2.3025851 7.5000000 -0.6931472 4.6051702
#>
#> Omega ($omega):
#> eta.ktr eta.ka eta.cl eta.v eta.emax eta.c50 eta.e0
#> eta.ktr 1 0 0 0 0.0 0.0 0.0
#> eta.ka 0 1 0 0 0.0 0.0 0.0
#> eta.cl 0 0 2 0 0.0 0.0 0.0
#> eta.v 0 0 0 1 0.0 0.0 0.0
#> eta.emax 0 0 0 0 0.5 0.0 0.0
#> eta.c50 0 0 0 0 0.0 0.5 0.0
#> eta.e0 0 0 0 0 0.0 0.0 0.5
#> -- Multiple Endpoint Model ($multipleEndpoint): ------------------------------------------------------------------------
#> variable cmt dvid*
#> 1 cp ~ ... cmt='center' or cmt=3 dvid='center' or dvid=1
#> 2 effect ~ ... cmt='ce' or cmt=4 dvid='ce' or dvid=2
#>
#> -- mu-referencing ($muRefTable): ---------------------------------------------------------------------------------------
#> theta eta
#> 1 tktr eta.ktr
#> 2 tka eta.ka
#> 3 tcl eta.cl
#> 4 tv eta.v
#> 5 poplogit eta.emax
#> 6 tc50 eta.c50
#> 7 te0 eta.e0
#>
#> -- Model: --------------------------------------------------------------------------------------------------------------
#> ktr <- exp(tktr + eta.ktr)
#> ka <- exp(tka + eta.ka)
#> cl <- exp(tcl + eta.cl)
#> v <- exp(tv + eta.v)
#>
#> logit=poplogit+eta.emax
#> emax = 1/(1+exp(-logit))
#> c50 = exp(tc50 + eta.c50)
#> e0 = exp(te0 + eta.e0)
#>
#> cp = center / v
#> d/dt(depot) = -ktr * depot
#> d/dt(gut) = ktr * depot -ka * gut
#> d/dt(center) = ka * gut - cl * cp
#>
#> effect=e0*(1-emax*cp/(c50+cp))
#>
#> cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
#> effect ~ add(eps.pdadd) | ce
#> ________________________________________________________________________________________________________________________
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [1] "CMT"
#> -- nlmixr SAEM(ODE); OBJF by FOCEi approximation fit -------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 2634.103 3555.798 3626.858 -1760.899 817643.9
#>
#> -- Time (sec pkpd.fit.CPS$time): ---------------------------------------------------------------------------------------
#>
#> saem setup table cwres covariance other
#> elapsed 70.96 55.52 0.27 56 0.07 3.97
#>
#> -- Population Parameters (pkpd.fit.CPS$parFixed or pkpd.fit.CPS$parFixedDf): -------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV% or SD)
#> tktr -1.77 1.14 64.1 0.17 (0.0183, 1.58) 432.
#> tka -2.72 0.768 28.3 0.0662 (0.0147, 0.298) 318.
#> tcl -3.46 1.1 31.7 0.0316 (0.00369, 0.27) 23.3
#> tv 2.93 0.192 6.56 18.7 (12.8, 27.3) 21.7
#> eps.pkprop 0.374 0.374
#> eps.pkadd 5.32 5.32
#> poplogit 7.16 72 1.01e+003 7.16 (-134, 148) 0.442
#> tc50 0.183 0.32 175 1.2 (0.641, 2.25) 49.3
#> te0 4.52 0.0821 1.82 91.5 (77.9, 107) 45.3
#> eps.pdadd 11.7 11.7
#> Shrink(SD)%
#> tktr 44.7%
#> tka 31.6%
#> tcl 86.8%
#> tv 60.7%
#> eps.pkprop
#> eps.pkadd
#> poplogit 90.0%
#> tc50 31.9%
#> te0 4.33%
#> eps.pdadd
#>
#> Covariance Type (pkpd.fit.CPS$covMethod): linFim
#> Some strong fixed parameter correlations exist (pkpd.fit.CPS$cor) :
#> cor:tka,tktr cor:tcl,tktr cor:tv,tktr cor:poplogit,tktr
#> -0.721 0.0907 -0.176 0.292
#> cor:tc50,tktr cor:te0,tktr cor:tcl,tka cor:tv,tka
#> 0.211 0.0139 -0.413 0.485
#> cor:poplogit,tka cor:tc50,tka cor:te0,tka cor:tv,tcl
#> -0.0435 0.0311 -0.00681 -0.668
#> cor:poplogit,tcl cor:tc50,tcl cor:te0,tcl cor:poplogit,tv
#> -0.0230 -0.133 0.000197 0.0964
#> cor:tc50,tv cor:te0,tv cor:tc50,poplogit cor:te0,poplogit
#> -0.0560 -0.000944 0.863 -0.0237
#> cor:te0,tc50
#> -0.0614
#>
#>
#> No correlations in between subject variability (BSV) matrix
#> Full BSV covariance (pkpd.fit.CPS$omega)
#> or correlation ( pkpd.fit.CPS $omegaR ; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#>
#> -- Fit Data (object pkpd.fit.CPS is a modified tibble): ----------------------------------------------------------------
#> # A tibble: 483 x 35
#> ID TIME CMT DV PRED RES WRES IPRED IRES IWRES CPRED
#> <fct> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.5 center 0.338 0.00721 0.331 0.0622 0.000495 0.338 0.0634 0.00183
#> 2 1 1 center 0.784 0.0277 0.757 0.142 0.00196 0.782 0.147 0.00722
#> 3 1 2 center 5.09 0.103 4.99 0.936 0.00769 5.08 0.955 0.0281
#> # ... with 480 more rows, and 24 more variables: CRES <dbl>, CWRES <dbl>,
#> # eta.ktr <dbl>, eta.ka <dbl>, eta.cl <dbl>, eta.v <dbl>, eta.emax <dbl>,
#> # eta.c50 <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>,
#> # ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>, logit <dbl>, emax <dbl>, c50 <dbl>,
#> # e0 <dbl>, cp <dbl>, effect <dbl>, tad <dbl>, dosenum <dbl>8.1.2 pkpd.fit.CPS VPC & GOF
vpc=myvpc(pkpd.fit.CPS,ylab="pkpd CPS Response (PCA) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:05
gof=mygof(pkpd.fit.CPS)
vpc/gof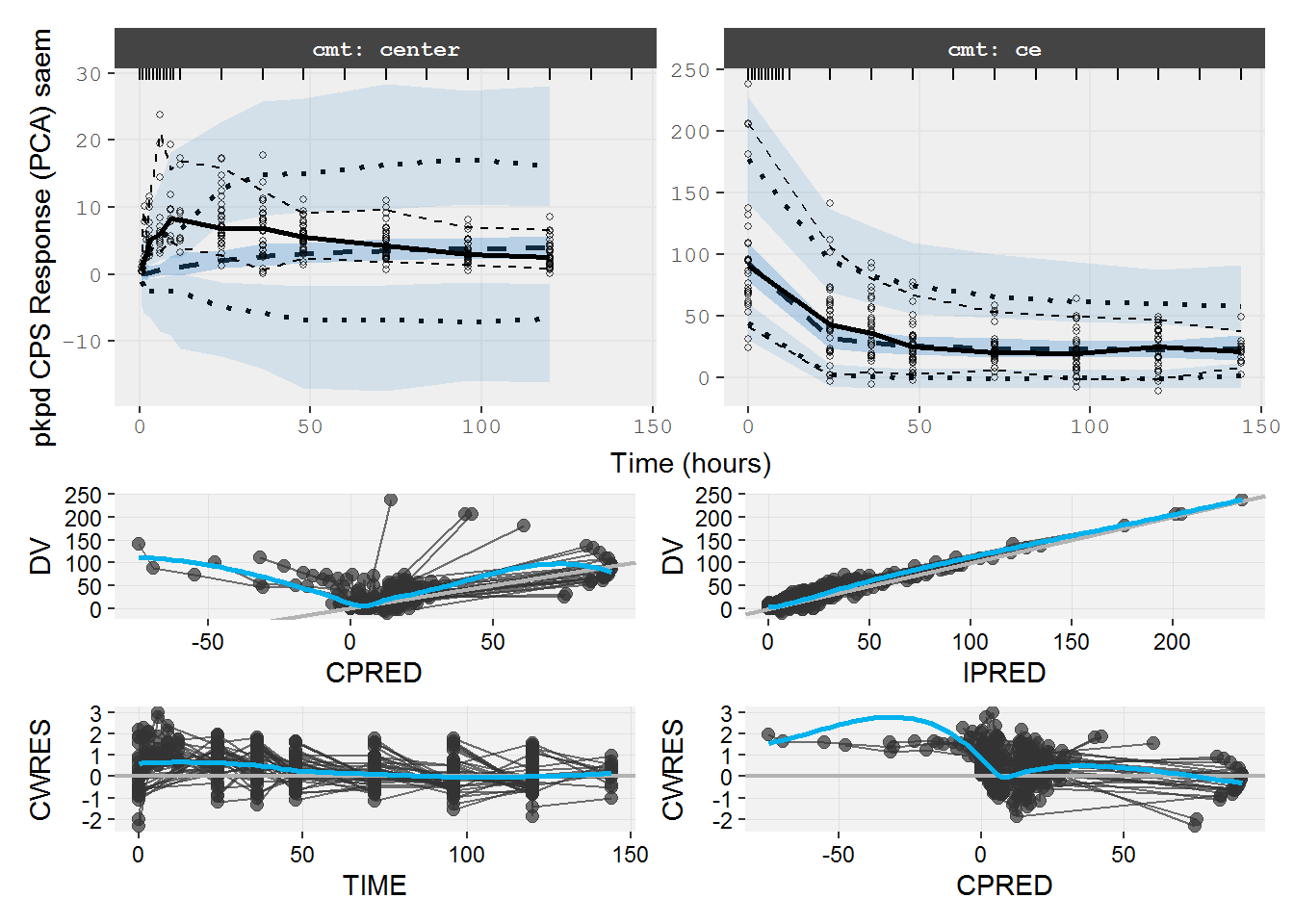
8.2 pkpd.ce.logit Effect CMT PKPD logit
Joint effect CMT logit
pkpd.ce.logit <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
eps.pkprop <- 0.1
eps.pkadd <- 0.1
poplogit <- 7.5
tc50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
cp = center / v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl * cp
d/dt(ce) = kout*(cp - ce)
effect=e0*(1-emax*ce/(c50+ce))
cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
effect ~ add(eps.pdadd) | ce
})
}8.2.1 pkpd.fit.CES SAEM
Effect compartment model using the combined PKPD data.
#> __ RxODE-based ODE model _______________________________________________________________________________________________
#> -- Initialization: -----------------------------------------------------------------------------------------------------
#> Fixed Effects ($theta):
#> tktr tka tcl tv poplogit tc50 tkout
#> 0.0000000 0.0000000 -2.3025851 2.3025851 7.5000000 -0.6931472 -2.9957323
#> te0
#> 4.6051702
#>
#> Omega ($omega):
#> eta.ktr eta.ka eta.cl eta.v eta.emax eta.c50 eta.kout eta.e0
#> eta.ktr 1 0 0 0 0.0 0.0 0.0 0.0
#> eta.ka 0 1 0 0 0.0 0.0 0.0 0.0
#> eta.cl 0 0 2 0 0.0 0.0 0.0 0.0
#> eta.v 0 0 0 1 0.0 0.0 0.0 0.0
#> eta.emax 0 0 0 0 0.5 0.0 0.0 0.0
#> eta.c50 0 0 0 0 0.0 0.5 0.0 0.0
#> eta.kout 0 0 0 0 0.0 0.0 0.5 0.0
#> eta.e0 0 0 0 0 0.0 0.0 0.0 0.5
#> -- Multiple Endpoint Model ($multipleEndpoint): ------------------------------------------------------------------------
#> variable cmt dvid*
#> 1 cp ~ ... cmt='center' or cmt=3 dvid='center' or dvid=1
#> 2 effect ~ ... cmt='ce' or cmt=4 dvid='ce' or dvid=2
#>
#> -- mu-referencing ($muRefTable): ---------------------------------------------------------------------------------------
#> theta eta
#> 1 tktr eta.ktr
#> 2 tka eta.ka
#> 3 tcl eta.cl
#> 4 tv eta.v
#> 5 poplogit eta.emax
#> 6 tc50 eta.c50
#> 7 tkout eta.kout
#> 8 te0 eta.e0
#>
#> -- Model: --------------------------------------------------------------------------------------------------------------
#> ktr <- exp(tktr + eta.ktr)
#> ka <- exp(tka + eta.ka)
#> cl <- exp(tcl + eta.cl)
#> v <- exp(tv + eta.v)
#>
#> logit=poplogit+eta.emax
#> emax = 1/(1+exp(-logit))
#> c50 = exp(tc50 + eta.c50)
#> kout = exp(tkout + eta.kout)
#> e0 = exp(te0 + eta.e0)
#>
#> cp = center / v
#> d/dt(depot) = -ktr * depot
#> d/dt(gut) = ktr * depot -ka * gut
#> d/dt(center) = ka * gut - cl * cp
#>
#> d/dt(ce) = kout*(cp - ce)
#>
#> effect=e0*(1-emax*ce/(c50+ce))
#>
#> cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
#> effect ~ add(eps.pdadd) | ce
#> ________________________________________________________________________________________________________________________
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [1] "CMT"
#> -- nlmixr SAEM(ODE); OBJF by FOCEi approximation fit -------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 2204.67 3130.364 3209.785 -1546.182 144712.4
#>
#> -- Time (sec pkpd.fit.CES$time): ---------------------------------------------------------------------------------------
#>
#> saem setup table cwres covariance other
#> elapsed 143.11 52.095 0.36 52.8 0.08 2.475
#>
#> -- Population Parameters (pkpd.fit.CES$parFixed or pkpd.fit.CES$parFixedDf): -------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV% or SD)
#> tktr 0.675 0.594 88.1 1.96 (0.613, 6.29) 133.
#> tka -0.735 0.254 34.5 0.479 (0.291, 0.788) 28.4
#> tcl -2.2 0.109 4.96 0.111 (0.0892, 0.137) 48.2
#> tv 2.49 0.0838 3.36 12.1 (10.2, 14.2) 32.9
#> eps.pkprop 0.538 0.538
#> eps.pkadd 0.0149 0.0149
#> poplogit 5.92 25.8 435 5.92 (-44.6, 56.5) 0.810
#> tc50 -0.617 0.864 140 0.54 (0.0992, 2.93) 22.9
#> tkout -5.32 0.772 14.5 0.00488 (0.00107, 0.0222) 40.3
#> te0 4.52 0.0824 1.82 91.7 (78, 108) 45.7
#> eps.pdadd 12.4 12.4
#> Shrink(SD)%
#> tktr 75.5%
#> tka 79.7%
#> tcl 16.7%
#> tv 22.9%
#> eps.pkprop
#> eps.pkadd
#> poplogit 89.3%
#> tc50 59.1%
#> tkout 43.0%
#> te0 3.20%
#> eps.pdadd
#>
#> Covariance Type (pkpd.fit.CES$covMethod): linFim
#> Some strong fixed parameter correlations exist (pkpd.fit.CES$cor) :
#> cor:tka,tktr cor:tcl,tktr cor:tv,tktr cor:poplogit,tktr
#> -0.638 -0.0355 0.0155 -0.0476
#> cor:tc50,tktr cor:tkout,tktr cor:te0,tktr cor:tcl,tka
#> -0.0385 -0.0288 0.00243 -0.0526
#> cor:tv,tka cor:poplogit,tka cor:tc50,tka cor:tkout,tka
#> 0.118 0.0575 0.0481 0.0375
#> cor:te0,tka cor:tv,tcl cor:poplogit,tcl cor:tc50,tcl
#> -0.00459 -0.153 0.0206 -0.0631
#> cor:tkout,tcl cor:te0,tcl cor:poplogit,tv cor:tc50,tv
#> -0.0887 -0.00132 0.00557 0.0463
#> cor:tkout,tv cor:te0,tv cor:tc50,poplogit cor:tkout,poplogit
#> 0.103 0.00255 0.892 0.795
#> cor:te0,poplogit cor:tkout,tc50 cor:te0,tc50 cor:te0,tkout
#> -0.0214 0.973 -0.0198 0.00459
#>
#>
#> No correlations in between subject variability (BSV) matrix
#> Full BSV covariance (pkpd.fit.CES$omega)
#> or correlation ( pkpd.fit.CES $omegaR ; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#>
#> -- Fit Data (object pkpd.fit.CES is a modified tibble): ----------------------------------------------------------------
#> # A tibble: 483 x 38
#> ID TIME CMT DV PRED RES WRES IPRED IRES IWRES CPRED CRES
#> <fct> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.5 center 0.338 0.661 -0.323 -0.493 0.479 -0.140 -0.544 0.639 -0.301
#> 2 1 1 center 0.784 1.87 -1.08 -0.688 1.44 -0.656 -0.847 1.84 -1.06
#> 3 1 2 center 5.09 4.10 0.990 0.344 3.45 1.64 0.887 4.12 0.975
#> # ... with 480 more rows, and 26 more variables: CWRES <dbl>, eta.ktr <dbl>,
#> # eta.ka <dbl>, eta.cl <dbl>, eta.v <dbl>, eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.kout <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>,
#> # ce <dbl>, ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>, logit <dbl>, emax <dbl>,
#> # c50 <dbl>, kout <dbl>, e0 <dbl>, cp <dbl>, effect <dbl>, tad <dbl>,
#> # dosenum <dbl>8.2.2 pkpd.fit.CES VPC & GOF
VPC medians do not agree. Note the time trends in the GOF plots (e.g. CPRED vs CWRES).
vpc=myvpc(pkpd.fit.CES,ylab="pkpd CES Response (PCA) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:08
gof=mygof(pkpd.fit.CES)
vpc/gof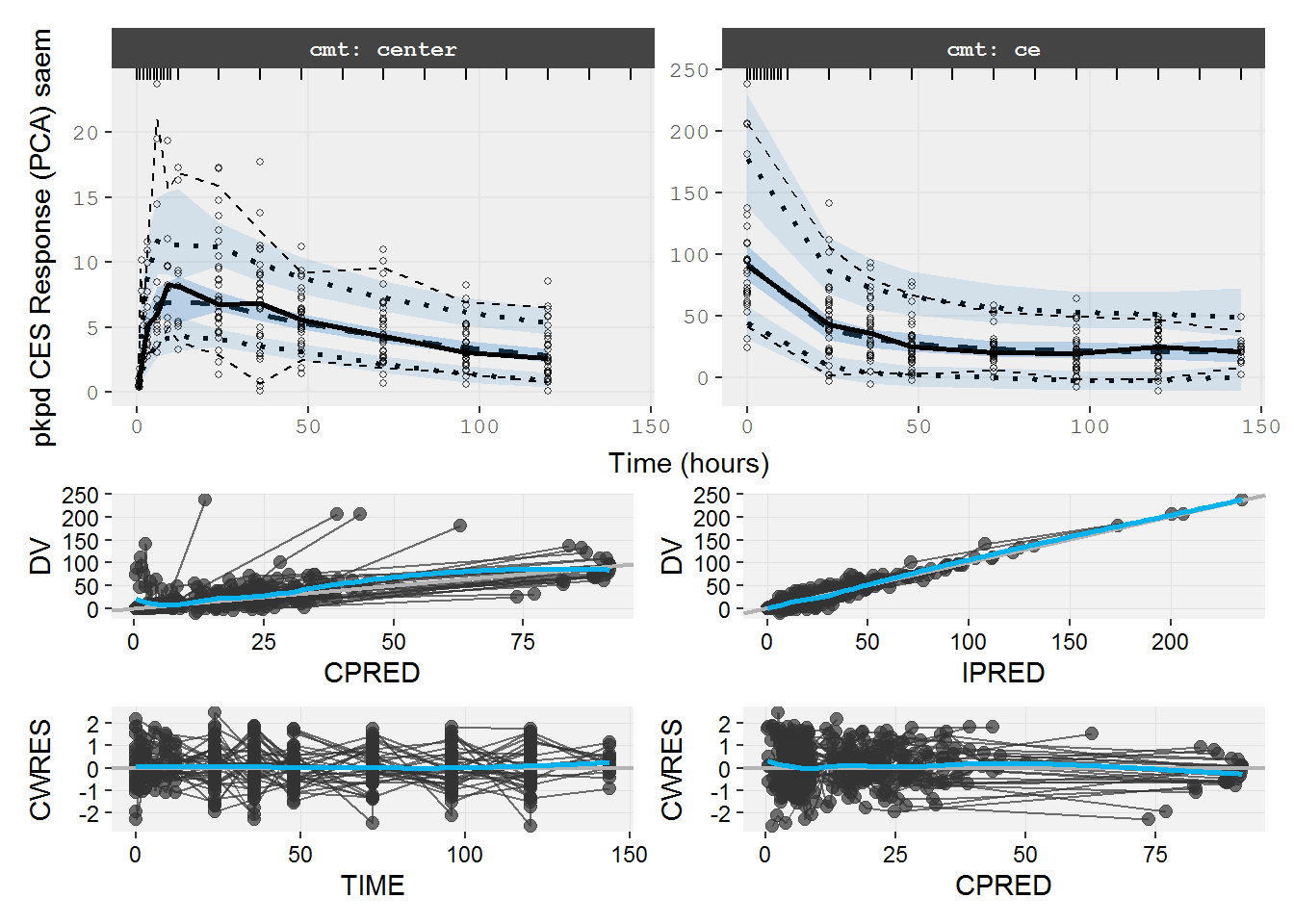
8.3 pkpd.turnover.logit TO PKPD logit
Joint turnover model logit
pkpd.turnover.logit <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
eps.pkprop <- 0.1
eps.pkadd <- 0.1
poplogit <- 7.5
tc50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
logit=poplogit+eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
cp = center / v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl * cp
effect(0) = e0
kin = e0*kout
PD=1-emax*cp/(c50+cp)
d/dt(effect) = kin*PD -kout*effect
cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
effect ~ add(eps.pdadd) | effect
})
}8.3.1 pkpd.fit.TOS SAEM
################ Turnover (true) PKPD logit model SAEM ###########
nlmixr(pkpd.turnover.logit) # Show initial estimates and model
#> __ RxODE-based ODE model _______________________________________________________________________________________________
#> -- Initialization: -----------------------------------------------------------------------------------------------------
#> Fixed Effects ($theta):
#> tktr tka tcl tv poplogit tc50 tkout
#> 0.0000000 0.0000000 -2.3025851 2.3025851 7.5000000 -0.6931472 -2.9957323
#> te0
#> 4.6051702
#>
#> Omega ($omega):
#> eta.ktr eta.ka eta.cl eta.v eta.emax eta.c50 eta.kout eta.e0
#> eta.ktr 1 0 0 0 0.0 0.0 0.0 0.0
#> eta.ka 0 1 0 0 0.0 0.0 0.0 0.0
#> eta.cl 0 0 2 0 0.0 0.0 0.0 0.0
#> eta.v 0 0 0 1 0.0 0.0 0.0 0.0
#> eta.emax 0 0 0 0 0.5 0.0 0.0 0.0
#> eta.c50 0 0 0 0 0.0 0.5 0.0 0.0
#> eta.kout 0 0 0 0 0.0 0.0 0.5 0.0
#> eta.e0 0 0 0 0 0.0 0.0 0.0 0.5
#> -- Multiple Endpoint Model ($multipleEndpoint): ------------------------------------------------------------------------
#> variable cmt dvid*
#> 1 cp ~ ... cmt='center' or cmt=3 dvid='center' or dvid=1
#> 2 effect ~ ... cmt='effect' or cmt=4 dvid='effect' or dvid=2
#>
#> -- mu-referencing ($muRefTable): ---------------------------------------------------------------------------------------
#> theta eta
#> 1 tktr eta.ktr
#> 2 tka eta.ka
#> 3 tcl eta.cl
#> 4 tv eta.v
#> 5 poplogit eta.emax
#> 6 tc50 eta.c50
#> 7 tkout eta.kout
#> 8 te0 eta.e0
#>
#> -- Model: --------------------------------------------------------------------------------------------------------------
#> ktr <- exp(tktr + eta.ktr)
#> ka <- exp(tka + eta.ka)
#> cl <- exp(tcl + eta.cl)
#> v <- exp(tv + eta.v)
#>
#> logit=poplogit+eta.emax
#> emax = 1/(1+exp(-logit))
#> c50 = exp(tc50 + eta.c50)
#> kout = exp(tkout + eta.kout)
#> e0 = exp(te0 + eta.e0)
#>
#> cp = center / v
#> d/dt(depot) = -ktr * depot
#> d/dt(gut) = ktr * depot -ka * gut
#> d/dt(center) = ka * gut - cl * cp
#>
#> effect(0) = e0
#> kin = e0*kout
#> PD=1-emax*cp/(c50+cp)
#> d/dt(effect) = kin*PD -kout*effect
#>
#> cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
#> effect ~ add(eps.pdadd) | effect
#> ________________________________________________________________________________________________________________________
pkpd.fit.TOS <- nlmixr(pkpd.turnover.logit, data.pkpd, est="saem", control=list(print=0),table=list(cwres=T))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [1] "CMT"
if (isplot) plot(pkpd.fit.TOS)
pkpd.fit.TOS
#> -- nlmixr SAEM(ODE); OBJF by FOCEi approximation fit -------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 2327.036 3252.73 3332.151 -1607.365 404302.4
#>
#> -- Time (sec pkpd.fit.TOS$time): ---------------------------------------------------------------------------------------
#>
#> saem setup table cwres covariance other
#> elapsed 151.49 65.466 0.36 66.22 0.22 2.464
#>
#> -- Population Parameters (pkpd.fit.TOS$parFixed or pkpd.fit.TOS$parFixedDf): -------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV% or SD)
#> tktr -0.381 0.166 43.5 0.683 (0.493, 0.945) 33.7
#> tka -0.01 0.0994 990 0.99 (0.815, 1.2) 20.5
#> tcl -1.84 0.137 7.44 0.158 (0.121, 0.207) 19.5
#> tv 2.64 0.123 4.67 14.1 (11, 17.9) 29.1
#> eps.pkprop 0.482 0.482
#> eps.pkadd 2.93 2.93
#> poplogit 7.06 52.6 746 7.06 (-96.1, 110) 0.862
#> tc50 -0.572 0.338 59.1 0.564 (0.291, 1.09) 65.9
#> tkout -3.29 0.118 3.59 0.0373 (0.0296, 0.047) 49.7
#> te0 4.51 0.084 1.86 90.9 (77.1, 107) 47.6
#> eps.pdadd 10.7 10.7
#> Shrink(SD)%
#> tktr 87.5%
#> tka 89.1%
#> tcl 60.3%
#> tv 48.0%
#> eps.pkprop
#> eps.pkadd
#> poplogit 90.6%
#> tc50 29.0%
#> tkout 19.2%
#> te0 2.40%
#> eps.pdadd
#>
#> Covariance Type (pkpd.fit.TOS$covMethod): linFim
#> Some strong fixed parameter correlations exist (pkpd.fit.TOS$cor) :
#> cor:tka,tktr cor:tcl,tktr cor:tv,tktr cor:poplogit,tktr
#> -0.0440 -0.0108 0.0605 -0.118
#> cor:tc50,tktr cor:tkout,tktr cor:te0,tktr cor:tcl,tka
#> -0.0865 0.0459 0.00561 -0.000269
#> cor:tv,tka cor:poplogit,tka cor:tc50,tka cor:tkout,tka
#> 0.0283 0.00227 -0.0133 -0.0142
#> cor:te0,tka cor:tv,tcl cor:poplogit,tcl cor:tc50,tcl
#> -0.000736 -0.168 -0.0612 -0.439
#> cor:tkout,tcl cor:te0,tcl cor:poplogit,tv cor:tc50,tv
#> -0.218 -0.0108 0.0909 0.147
#> cor:tkout,tv cor:te0,tv cor:tc50,poplogit cor:tkout,poplogit
#> 0.165 0.00733 0.792 -0.402
#> cor:te0,poplogit cor:tkout,tc50 cor:te0,tc50 cor:te0,tkout
#> -0.0299 -0.156 -0.0379 0.0804
#>
#>
#> No correlations in between subject variability (BSV) matrix
#> Full BSV covariance (pkpd.fit.TOS$omega)
#> or correlation ( pkpd.fit.TOS $omegaR ; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#>
#> -- Fit Data (object pkpd.fit.TOS is a modified tibble): ----------------------------------------------------------------
#> # A tibble: 483 x 39
#> ID TIME CMT DV PRED RES WRES IPRED IRES IWRES CPRED
#> <fct> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.5 center 0.338 0.456 -0.118 -0.0401 0.405 -0.0664 -0.0226 0.453
#> 2 1 1 center 0.784 1.40 -0.616 -0.202 1.25 -0.465 -0.155 1.39
#> 3 1 2 center 5.09 3.41 1.68 0.470 3.07 2.02 0.614 3.40
#> # ... with 480 more rows, and 28 more variables: CRES <dbl>, CWRES <dbl>,
#> # eta.ktr <dbl>, eta.ka <dbl>, eta.cl <dbl>, eta.v <dbl>, eta.emax <dbl>,
#> # eta.c50 <dbl>, eta.kout <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>,
#> # center <dbl>, effect <dbl>, ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>,
#> # logit <dbl>, emax <dbl>, c50 <dbl>, kout <dbl>, e0 <dbl>, cp <dbl>,
#> # kin <dbl>, PD <dbl>, tad <dbl>, dosenum <dbl>8.3.2 pkpd.fit.TOS VPC
VPCs look acceptable and no problem visible in the standard GOF plots.
vpc=myvpc(pkpd.fit.TOS,ylab="pkpd TOS Response (PCA) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:03
gof=mygof(pkpd.fit.TOS)
vpc/gof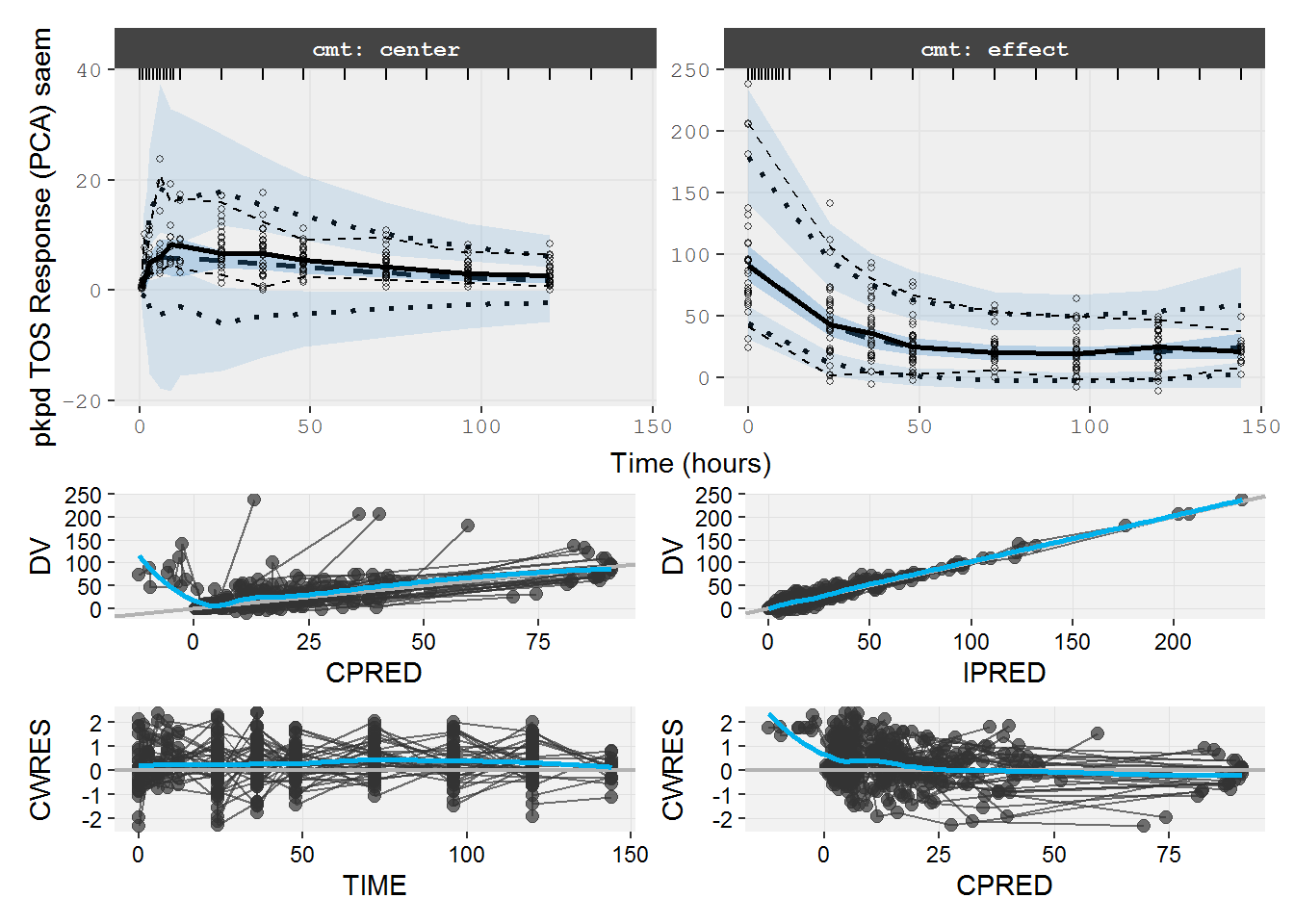
8.4 pkpd.cp.emax Imm Effect PKPD Emax
Joint immediate effect model Emax<1
pkpd.cp.emax <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
eps.pkprop <- 0.1
eps.pkadd <- 0.1
popemax <- c(0,0.9,0.999)
tc50 <- log(0.5)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
poplogit = log(popemax/(1-popemax))
logit=poplogit + eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
e0 = exp(te0 + eta.e0)
cp = center / v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl * cp
effect=e0*(1-emax*cp/(c50+cp))
cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
effect ~ add(eps.pdadd) | effect
})
}8.4.1 pkpd.fit.CPF FOCEI
################ Immediate PKPD emax model FOCEI ###########
pkpd.fit.CPF <- nlmixr(pkpd.cp.emax, data.pkpd, est="focei", control=list(print=0,outerOpt="bobyqa",table=list(cwres=T)))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:03
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "CMT"
#> unhandled error message: EE:lsoda -- at t = 0.00198818 and step size _C(h) = 7.58429e-009, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at start of problem, too much accuracy
#> requested for precision of machine,
#> suggested scaling factor = 431.47
#> @(lsoda.c:761)
#> unhandled error message: EE:lsoda -- at start of problem, too much accuracy
#> requested for precision of machine,
#> suggested scaling factor = 430.365
#> @(lsoda.c:761)
#> unhandled error message: EE:lsoda -- at start of problem, too much accuracy
#> requested for precision of machine,
#> suggested scaling factor = 64.5623
#> @(lsoda.c:761)
#> unhandled error message: EE:lsoda -- at start of problem, too much accuracy
#> requested for precision of machine,
#> suggested scaling factor = 426.656
#> @(lsoda.c:761)
#> unhandled error message: EE:lsoda -- at start of problem, too much accuracy
#> requested for precision of machine,
#> suggested scaling factor = 368.143
#> @(lsoda.c:761)
#> unhandled error message: EE:lsoda -- at start of problem, too much accuracy
#> requested for precision of machine,
#> suggested scaling factor = 162.742
#> @(lsoda.c:761)
#> unhandled error message: EE:lsoda -- at start of problem, too much accuracy
#> requested for precision of machine,
#> suggested scaling factor = 402.316
#> @(lsoda.c:761)ll
#> unhandled error message: EE:lsoda -- at start of problem, too much accuracy
#> requested for precision of machine,
#> suggested scaling factor = 415.77
#> @(lsoda.c:761)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.97483e-009, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 7.58425e-009, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> Error in .fitFun(.ret) :
#> Evaluation error: Starting values violate bounds.
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:01:56
#> done
if (isplot) plot(pkpd.fit.CPF)
pkpd.fit.CPF
#> -- nlmixr FOCEi (outer: bobyqa) fit ------------------------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 2269.373 3191.068 3262.128 -1578.534 16115.44
#>
#> -- Time (sec pkpd.fit.CPF$time): ---------------------------------------------------------------------------------------
#>
#> setup optimize covariance table other
#> elapsed 82.679 116.414 116.414 0.17 524.283
#>
#> -- Population Parameters (pkpd.fit.CPF$parFixed or pkpd.fit.CPF$parFixedDf): -------------------------------------------
#>
#> Est. SE %RSE Back-transformed(95%CI) BSV(CV%)
#> tktr -1.26 0.327 25.9 0.283 (0.149, 0.538) 11.9
#> tka 2.28 1.76 77 9.83 (0.312, 309) 39.3
#> tcl -2.17 0.184 8.5 0.114 (0.0795, 0.164) 43.5
#> tv 2.34 0.185 7.89 10.4 (7.24, 14.9) 45.4
#> eps.pkprop 0.317 0.317
#> eps.pkadd 0.000289 0.000289
#> popemax 0.706 0.0537 7.6 0.706 (0.601, 0.812)
#> tc50 -3.5 5.76 164 0.0302 (3.8e-007, 2.4e+003) 323.
#> te0 4.5 0.148 3.29 90.2 (67.5, 121) 41.2
#> eps.pdadd 17.5 17.5
#> Shrink(SD)%
#> tktr 78.5%
#> tka 86.7%
#> tcl 13.1%
#> tv 7.50%
#> eps.pkprop
#> eps.pkadd
#> popemax
#> tc50 90.9%
#> te0 6.30%
#> eps.pdadd
#>
#> -- BSV Covariance (pkpd.fit.CPF$omega): --------------------------------------------------------------------------------
#> eta.ktr eta.ka eta.cl eta.v eta.emax eta.c50 eta.e0
#> eta.ktr 0.01414784 0.0000000 0.0000000 0.0000000 0.000000 0.000000 0.000000
#> eta.ka 0.00000000 0.1434983 0.0000000 0.0000000 0.000000 0.000000 0.000000
#> eta.cl 0.00000000 0.0000000 0.1732686 0.0000000 0.000000 0.000000 0.000000
#> eta.v 0.00000000 0.0000000 0.0000000 0.1873769 0.000000 0.000000 0.000000
#> eta.emax 0.00000000 0.0000000 0.0000000 0.0000000 0.179734 0.000000 0.000000
#> eta.c50 0.00000000 0.0000000 0.0000000 0.0000000 0.000000 2.433876 0.000000
#> eta.e0 0.00000000 0.0000000 0.0000000 0.0000000 0.000000 0.000000 0.156704
#>
#> Not all variables are mu-referenced.
#> Can also see BSV Correlation (pkpd.fit.CPF$omegaR; diagonals=SDs)
#> Covariance Type (pkpd.fit.CPF$covMethod): s
#> Some strong fixed parameter correlations exist (pkpd.fit.CPF$cor) :
#> cor:tka,tktr cor:tcl,tktr cor:tv,tktr cor:popemax,tktr
#> -0.726 0.262 0.307 -0.250
#> cor:tc50,tktr cor:te0,tktr cor:tcl,tka cor:tv,tka
#> 0.441 0.0688 -0.191 -0.155
#> cor:popemax,tka cor:tc50,tka cor:te0,tka cor:tv,tcl
#> 0.0237 -0.516 -0.0349 0.128
#> cor:popemax,tcl cor:tc50,tcl cor:te0,tcl cor:popemax,tv
#> 0.151 0.439 0.298 -0.116
#> cor:tc50,tv cor:te0,tv cor:tc50,popemax cor:te0,popemax
#> 0.131 -0.144 0.262 0.156
#> cor:te0,tc50
#> 0.221
#>
#>
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#> Minimization message (pkpd.fit.CPF$message):
#> Normal exit from bobyqa
#>
#> -- Fit Data (object pkpd.fit.CPF is a modified tibble): ----------------------------------------------------------------
#> # A tibble: 483 x 36
#> ID TIME CMT DV PRED RES WRES IPRED IRES IWRES CPRED CRES
#> <fct> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.5 center 0.338 1.02 -0.684 -1.21 0.987 -0.649 -2.08 1.02 -0.687
#> 2 1 1 center 0.784 2.15 -1.36 -1.16 2.12 -1.33 -1.99 2.15 -1.37
#> 3 1 2 center 5.09 3.95 1.14 0.532 3.94 1.15 0.921 3.96 1.13
#> # ... with 480 more rows, and 24 more variables: CWRES <dbl>, eta.ktr <dbl>,
#> # eta.ka <dbl>, eta.cl <dbl>, eta.v <dbl>, eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>, ktr <dbl>, ka <dbl>,
#> # cl <dbl>, v <dbl>, poplogit <dbl>, logit <dbl>, emax <dbl>, c50 <dbl>,
#> # e0 <dbl>, cp <dbl>, effect <dbl>, tad <dbl>, dosenum <dbl>8.4.2 pkpd.fit.CPF VPC & GOF
Compiling VPC model…done done (14.89 sec) Quitting from lines 1036-1043 (warf_simtr2_PKPD_vpc_ui_saem_foce_B-18-v3_PDF.Rmd) Error in as.character.factor(df[[col]]) : malformed factor Calls:
#vpc=myvpc(pkpd.fit.CPF,ylab="pkpd CPF Response (PCA) saem")
#gof=mygof(pkpd.fit.CPF)
#vpc/gof8.5 pkpd.ce.emax Effect CMT PKPD Emax
Joint effect compartment model Emax<1
pkpd.ce.emax <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
eps.pkprop <- 0.1
eps.pkadd <- 0.1
popemax <- c(0,0.9,0.999)
tc50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
poplogit = log(popemax/(1-popemax))
logit=poplogit + eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
cp = center / v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl * cp
d/dt(ce) = kout*(cp - ce)
effect=e0*(1-emax*ce/(c50+ce))
cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
effect ~ add(eps.pdadd) | ce
})
}8.5.1 pkpd.fit.CEF FOCEI
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "CMT"
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)16e
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)16e
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)16e
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)16e
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)16e
#> intdy -- t = 24 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 36 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 48 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 72 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 96 illegal. t not in interval tcur - _C(hu) to tcur
#> intdy -- t = 120 illegal. t not in interval tcur - _C(hu) to tcur
#> unhandled error message: EE:[lsoda] trouble from intdy, itask = 1, tout = 120
#> @(lsoda.c:689)
#> [====|====|====|====|====|====|====|====|====|====] 0:09:08
#> done
#> -- nlmixr FOCEi (outer: bobyqa) fit ------------------------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood
#> FOCEi 2136.444 3062.138 3141.559 -1512.069
#>
#> -- Time (sec pkpd.fit.CEF$time): ---------------------------------------------------------------------------------------
#>
#> setup optimize covariance table other
#> elapsed 72.23 0 0 0.12 553.66
#>
#> -- Population Parameters (pkpd.fit.CEF$parFixed or pkpd.fit.CEF$parFixedDf): -------------------------------------------
#>
#> Est. Back-transformed BSV(CV%) Shrink(SD)%
#> tktr 0.496 1.64 25.7 88.2%
#> tka -0.834 0.434 36.9 67.3%
#> tcl -2.19 0.111 44.0 10.8%
#> tv 2.41 11.1 41.9 12.1%
#> eps.pkprop 0.344 0.344
#> eps.pkadd 0.135 0.135
#> popemax 0.999 0.999
#> tc50 -0.782 0.457 22.0 59.8%
#> tkout -5.61 0.00367 38.7 39.9%
#> te0 4.53 93.2 44.7 5.10%
#> eps.pdadd 12.5 12.5
#>
#> -- BSV Covariance (pkpd.fit.CEF$omega): --------------------------------------------------------------------------------
#> eta.ktr eta.ka eta.cl eta.v eta.emax eta.c50 eta.kout
#> eta.ktr 0.06388799 0.0000000 0.0000000 0.0000000 0.00000 0.00000000 0.0000000
#> eta.ka 0.00000000 0.1279681 0.0000000 0.0000000 0.00000 0.00000000 0.0000000
#> eta.cl 0.00000000 0.0000000 0.1772143 0.0000000 0.00000 0.00000000 0.0000000
#> eta.v 0.00000000 0.0000000 0.0000000 0.1615624 0.00000 0.00000000 0.0000000
#> eta.emax 0.00000000 0.0000000 0.0000000 0.0000000 2.21256 0.00000000 0.0000000
#> eta.c50 0.00000000 0.0000000 0.0000000 0.0000000 0.00000 0.04745568 0.0000000
#> eta.kout 0.00000000 0.0000000 0.0000000 0.0000000 0.00000 0.00000000 0.1398162
#> eta.e0 0.00000000 0.0000000 0.0000000 0.0000000 0.00000 0.00000000 0.0000000
#> eta.e0
#> eta.ktr 0.0000000
#> eta.ka 0.0000000
#> eta.cl 0.0000000
#> eta.v 0.0000000
#> eta.emax 0.0000000
#> eta.c50 0.0000000
#> eta.kout 0.0000000
#> eta.e0 0.1824311
#>
#> Not all variables are mu-referenced.
#> Can also see BSV Correlation (pkpd.fit.CEF$omegaR; diagonals=SDs)
#> Covariance Type (pkpd.fit.CEF$covMethod): Boundary issue; Get SEs with getVarCov
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#> Minimization message (pkpd.fit.CEF$message):
#> Normal exit from bobyqa
#>
#> -- Fit Data (object pkpd.fit.CEF is a modified tibble): ----------------------------------------------------------------
#> # A tibble: 483 x 39
#> ID TIME CMT DV PRED RES WRES IPRED IRES IWRES CPRED CRES
#> <fct> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.5 center 0.338 0.574 -0.236 -0.592 0.510 -0.172 -0.775 0.571 -0.232
#> 2 1 1 center 0.784 1.69 -0.907 -0.850 1.53 -0.745 -1.37 1.68 -0.900
#> 3 1 2 center 5.09 3.94 1.15 0.494 3.65 1.44 1.14 3.94 1.15
#> # ... with 480 more rows, and 27 more variables: CWRES <dbl>, eta.ktr <dbl>,
#> # eta.ka <dbl>, eta.cl <dbl>, eta.v <dbl>, eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.kout <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>,
#> # ce <dbl>, ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>, poplogit <dbl>,
#> # logit <dbl>, emax <dbl>, c50 <dbl>, kout <dbl>, e0 <dbl>, cp <dbl>,
#> # effect <dbl>, tad <dbl>, dosenum <dbl>8.5.2 pkpd.fit.CEF VPC & GOF
vpc=myvpc(pkpd.fit.CEF,ylab="pkpd CEF Response (PCA) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:03
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "CMT"
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:00:07
#> [====|====|====|====|====|====|====|====|====|====
gof=mygof(pkpd.fit.CEF)
vpc/gof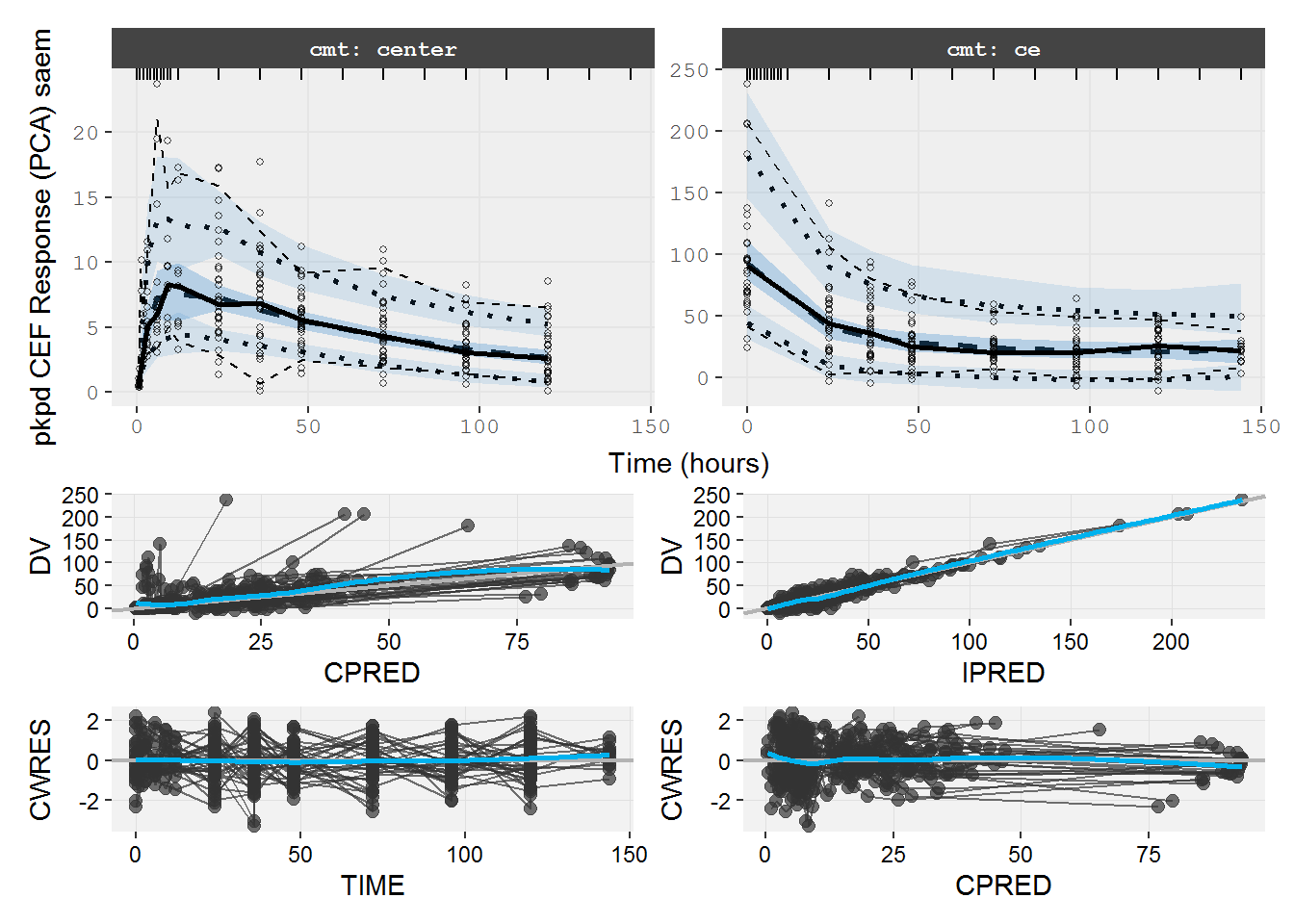
8.6 pkpd.turnover.emax TO PKPD Emax
Joint turnover model Emax<1
pkpd.turnover.emax <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
eps.pkprop <- 0.1
eps.pkadd <- 0.1
popemax <- c(0,0.9,0.999)
tc50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
eta.emax ~ .5
eta.c50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
eps.pdadd <- 100
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
poplogit = log(popemax/(1-popemax))
logit=poplogit + eta.emax
emax = 1/(1+exp(-logit))
c50 = exp(tc50 + eta.c50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
cp = center / v
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl * cp
effect(0) = e0
kin = e0*kout
PD=1-emax*cp/(c50+cp)
d/dt(effect) = kin*PD -kout*effect
cp ~ prop(eps.pkprop) + add(eps.pkadd) | center
effect ~ add(eps.pdadd) | effect
})
}8.6.1 pkpd.fit.TOF FOCEI
################ Turnover (true) PKPD emax model FOCEI ###########
pkpd.fit.TOF <- nlmixr(pkpd.turnover.emax, data.pkpd, est="focei", control=list(print=0,outerOpt="bobyqa",table=list(cwres=T)))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "CMT"
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 7.58746e-012, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> calculating covariance matrix
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 1.13456e-014, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47375e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 5.52427e-005 and step size _C(h) = 2.10734e-010, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 4.70256e-013, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0.579693 and step size _C(h) = 2.04951e-006, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)…Nö=
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 2.10734e-010, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 60.2022 and step size _C(h) = 3.27922e-005, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 6.03157e-008 and step size _C(h) = 4.49387e-016, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 8.61749 and step size _C(h) = 3.27922e-005, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 1.05367e-010, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 60.1951 and step size _C(h) = 3.27922e-005, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0.00707107 and step size _C(h) = 3.27922e-005, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887).
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 7.46815e-015, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 1.05367e-010, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 5.52794e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 1.05367e-010, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47375e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 1.05367e-010, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47375e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887).c:887)…Nö=
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47375e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 6.47376e-007, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> unhandled error message: EE:lsoda -- at t = 0 and step size _C(h) = 1.05367e-010, the
#> corrector convergence failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:887)
#> [====|====|====|====|====|====|====|====|====|=
S matrix calculation failed; Switch to R-matrix covariance.
#> ===] 0:08:13
#> done
if (isplot) plot(pkpd.fit.TOF)
pkpd.fit.TOF
#> -- nlmixr FOCEi (outer: bobyqa) fit ------------------------------------------------------------------------------------
#>
#> OBJF AIC BIC Log-likelihood
#> FOCEi 1.811158e+15 1.811158e+15 1.811158e+15 -9.055788e+14
#>
#> -- Time (sec pkpd.fit.TOF$time): ---------------------------------------------------------------------------------------
#>
#> setup optimize covariance table other
#> elapsed 67.607 493.37 493.37 0.22 -389.507
#>
#> -- Population Parameters (pkpd.fit.TOF$parFixed or pkpd.fit.TOF$parFixedDf): -------------------------------------------
#>
#> Est. Back-transformed BSV(CV%) Shrink(SD)%
#> tktr -0.0187 0.981 113. 74.5%
#> tka -0.026 0.974 106. 73.8%
#> tcl -2.33 0.0976 113. 73.3%
#> tv 2.31 10 71.9 4.58%
#> eps.pkprop 0.14 0.14
#> eps.pkadd 0.116 0.116
#> popemax 0.854 0.854
#> tc50 -0.699 0.497 79.2 2.85%
#> tkout -2.98 0.0506 74.2 -627.%
#> te0 4.54 93.9 59.1 48.0%
#> eps.pdadd 47 47
#>
#> -- BSV Covariance (pkpd.fit.TOF$omega): --------------------------------------------------------------------------------
#> eta.ktr eta.ka eta.cl eta.v eta.emax eta.c50 eta.kout
#> eta.ktr 0.8246359 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
#> eta.ka 0.0000000 0.7518864 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
#> eta.cl 0.0000000 0.0000000 0.8201897 0.0000000 0.0000000 0.0000000 0.0000000
#> eta.v 0.0000000 0.0000000 0.0000000 0.4165251 0.0000000 0.0000000 0.0000000
#> eta.emax 0.0000000 0.0000000 0.0000000 0.0000000 0.4649877 0.0000000 0.0000000
#> eta.c50 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.4866589 0.0000000
#> eta.kout 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.4390234
#> eta.e0 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
#> eta.e0
#> eta.ktr 0.0000000
#> eta.ka 0.0000000
#> eta.cl 0.0000000
#> eta.v 0.0000000
#> eta.emax 0.0000000
#> eta.c50 0.0000000
#> eta.kout 0.0000000
#> eta.e0 0.2999657
#>
#> Not all variables are mu-referenced.
#> Can also see BSV Correlation (pkpd.fit.TOF$omegaR; diagonals=SDs)
#> Covariance Type (pkpd.fit.TOF$covMethod): failed
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#> Minimization message (pkpd.fit.TOF$message):
#> Normal exit from bobyqa
#>
#> -- Fit Data (object pkpd.fit.TOF is a modified tibble): ----------------------------------------------------------------
#> # A tibble: 483 x 40
#> ID TIME CMT DV PRED RES WRES IPRED IRES IWRES CPRED CRES
#> <fct> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.5 center 0.338 0.863 -0.525 -0.485 0.870 -0.531 -3.16 0.863 -0.525
#> 2 1 1 center 0.784 2.54 -1.75 -0.628 2.56 -1.77 -4.71 2.54 -1.75
#> 3 1 2 center 5.09 5.74 -0.649 -0.129 5.77 -0.676 -0.829 5.74 -0.650
#> # ... with 480 more rows, and 28 more variables: CWRES <dbl>, eta.ktr <dbl>,
#> # eta.ka <dbl>, eta.cl <dbl>, eta.v <dbl>, eta.emax <dbl>, eta.c50 <dbl>,
#> # eta.kout <dbl>, eta.e0 <dbl>, depot <dbl>, gut <dbl>, center <dbl>,
#> # effect <dbl>, ktr <dbl>, ka <dbl>, cl <dbl>, v <dbl>, poplogit <dbl>,
#> # logit <dbl>, emax <dbl>, c50 <dbl>, kout <dbl>, e0 <dbl>, cp <dbl>,
#> # kin <dbl>, PD <dbl>, tad <dbl>, dosenum <dbl>8.6.2 pkpd.fit.TOF VPC
vpc=myvpc(pkpd.fit.TOF,ylab="pkpd TOF Response (PCA) saem")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "CMT"
#> calculating covariance matrix
#> unhandled error message: EE:lsoda -- at t = 105.035 and step size _C(h) = 1.39265e-008, the
#> error test failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:884)
#> [====|====|====|====|====|====|====|====|====|=
S matrix calculation failed; Switch to R-matrix covariance.
#> ===] 0:00:20
#> [1] "CMT"
#> calculating covariance matrix
#> unhandled error message: EE:lsoda -- at t = 105.035 and step size _C(h) = 1.39265e-008, the
#> error test failed repeatedly or
#> with fabs(_C(h)) = hmin
#> @(lsoda.c:884)
#> [====|====|====|====|====|====|====|====|====|=
S matrix calculation failed; Switch to R-matrix covariance.
#> ===] 0:00:20
#> [====|====|====|====|====|====|====|====|====|====
gof=mygof(pkpd.fit.TOF)
vpc/gof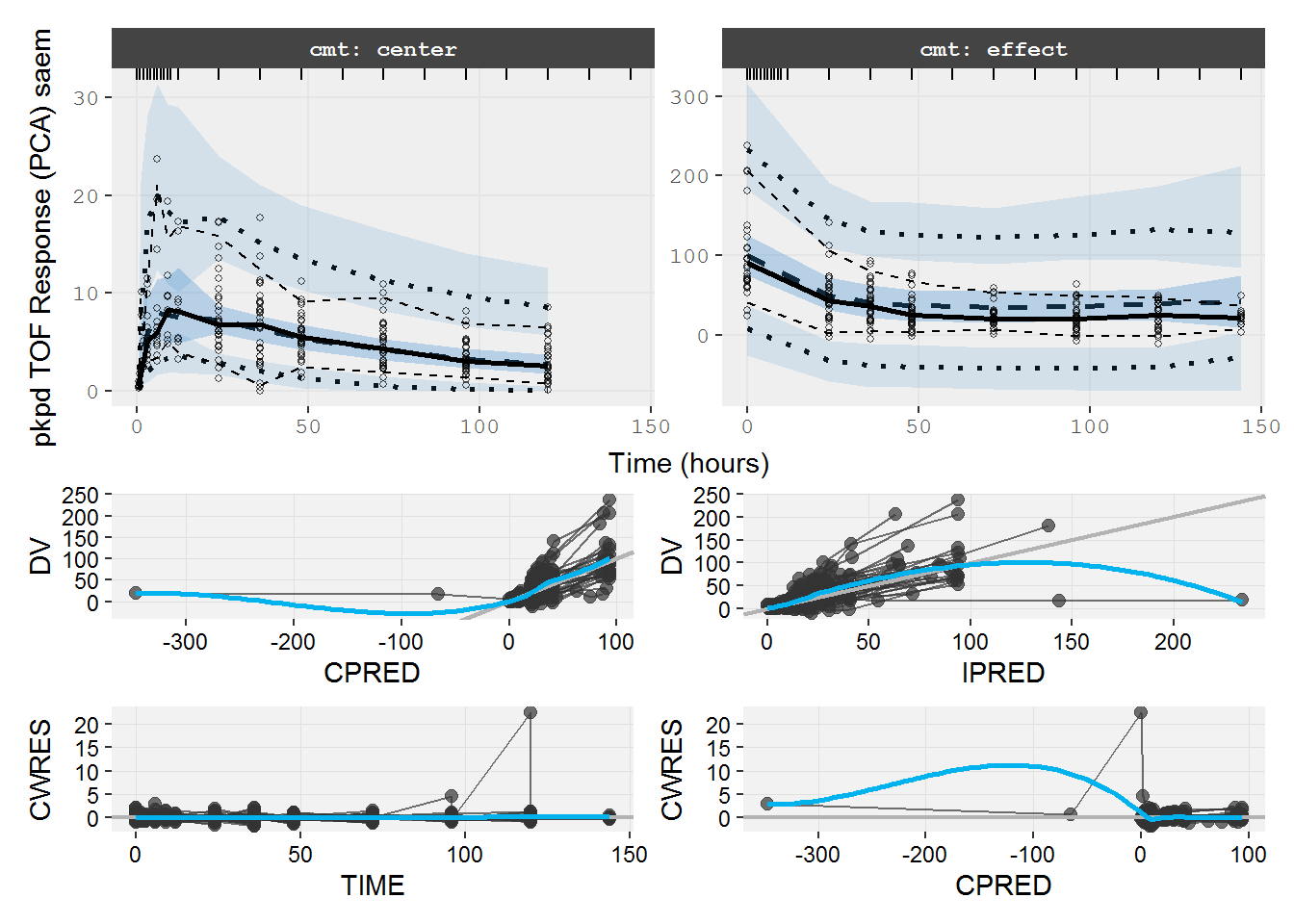
9 Rmarkdown
Output created: warf_simtr_PKPD_vpc_ui_saem_foce_B-18-v3_PDF.html